Enhanced Calculation of Property Distributions in Chemical Fragment Spaces
- PMID: 38466793
- PMCID: PMC10966640
- DOI: 10.1021/acs.jcim.4c00147
Enhanced Calculation of Property Distributions in Chemical Fragment Spaces
Abstract
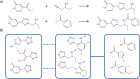
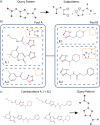
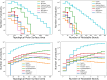
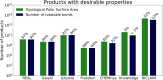
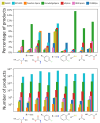
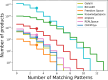
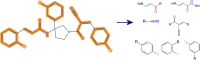
Chemical fragment spaces exceed traditional virtual compound libraries by orders of magnitude, making them ideal search spaces for drug design projects. However, due to their immense size, they are not compatible with traditional analysis and search algorithms that rely on the enumeration of molecules. In this paper, we present SpaceProp2, an evolution of the SpaceProp algorithm, which enables the calculation of exact property distributions for chemical fragment spaces without enumerating them. We extend the original algorithm by the capabilities to compute distributions for the TPSA, the number of rotatable bonds, and the occurrence of user-defined molecular structures in the form of SMARTS patterns. Furthermore, SpaceProp2 produces example molecules for every property bin, enabling a detailed interpretation of the distributions. We demonstrate SpaceProp2 on six established make-on-demand chemical fragment spaces as well as BICLAIM, the in-house fragment space of Boehringer Ingelheim. The possibility to search multiple SMARTS patterns simultaneously as well as the produced example molecules offers previously impossible insights into the composition of these vast combinatorial molecule collections, making it an ideal tool for the analysis and design of chemical fragment spaces.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors declare the following competing financial interest(s): M.R., as a shareholder of BioSolveIT GmbH, declares a potential financial interest in the event that the SpaceProp software is licensed for a fee to nonacademic institutions in the future.
Figures


References
-
- Enamine REAL Space. https://enamine.net/compound-collections/real-compounds/real-space-navig... (accessed June 26, 2023).
-
- GalaXi Space. https://www.labnetwork.com/frontend-app/p/#!/library/virtual (accessed June 26, 2023)
-
- CHEMriya Space. https://www.otavachemicals.com/products/chemriya (accessed June 26, 2023)
-
- eXplore Space. https://marketing.emolecules.com/explore (accessed June 26, 2023)
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

