Application of Mass Cytometry Platforms to Solid Organ Transplantation
- PMID: 38467594
- PMCID: PMC11390974
- DOI: 10.1097/TP.0000000000004925
Application of Mass Cytometry Platforms to Solid Organ Transplantation
Abstract
Transplantation serves as the cornerstone of treatment for patients with end-stage organ disease. The prevalence of complications, such as allograft rejection, infection, and malignancies, underscores the need to dissect the complex interactions of the immune system at the single-cell level. In this review, we discuss studies using mass cytometry or cytometry by time-of-flight, a cutting-edge technology enabling the characterization of immune populations and cell-to-cell interactions in granular detail. We review the application of mass cytometry in human and experimental animal studies in the context of transplantation, uncovering invaluable contributions of the tool to understanding rejection and other transplant-related complications. We discuss recent innovations that have the potential to streamline and standardize mass cytometry workflows for application to multisite clinical trials. Additionally, we introduce imaging mass cytometry, a technique that couples the power of mass cytometry with spatial context, thereby mapping cellular interactions within tissue microenvironments. The synergistic integration of mass cytometry and imaging mass cytometry data with other omics data sets and high-dimensional data platforms to further define immune dynamics is discussed. In conclusion, mass cytometry technologies, when integrated with other tools and data, shed light on the intricate landscape of the immune response in transplantation. This approach holds significant potential for enhancing patient outcomes by advancing our understanding and facilitating the development of new diagnostics and therapeutics.
Copyright © 2024 Wolters Kluwer Health, Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


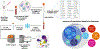
References
-
- Kim JV, Assadian S, Hollander Z, et al. Regulatory T cell biomarkers identify patients at risk of developing acute cellular rejection in the first year following heart transplantation. Transplantation 2023;107(8):1810–1819. - PubMed
-
- Angerer P, Haghverdi L, Buttner M, et al. Destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 2016;32(8):1241–1243. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

