Mitochondrial metabolism sustains CD8+ T cell migration for an efficient infiltration into solid tumors
- PMID: 38467616
- PMCID: PMC10928223
- DOI: 10.1038/s41467-024-46377-7
Mitochondrial metabolism sustains CD8+ T cell migration for an efficient infiltration into solid tumors
Abstract
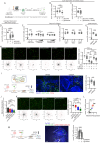
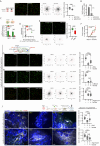
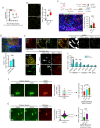
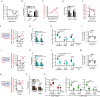
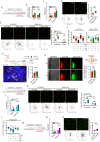
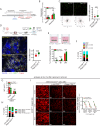
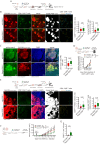
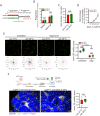
The ability of CD8+ T cells to infiltrate solid tumors and reach cancer cells is associated with improved patient survival and responses to immunotherapy. Thus, identifying the factors controlling T cell migration in tumors is critical, so that strategies to intervene on these targets can be developed. Although interstitial motility is a highly energy-demanding process, the metabolic requirements of CD8+ T cells migrating in a 3D environment remain unclear. Here, we demonstrate that the tricarboxylic acid (TCA) cycle is the main metabolic pathway sustaining human CD8+ T cell motility in 3D collagen gels and tumor slices while glycolysis plays a more minor role. Using pharmacological and genetic approaches, we report that CD8+ T cell migration depends on the mitochondrial oxidation of glucose and glutamine, but not fatty acids, and both ATP and ROS produced by mitochondria are required for T cells to migrate. Pharmacological interventions to increase mitochondrial activity improve CD8+ T cell intratumoral migration and CAR T cell recruitment into tumor islets leading to better control of tumor growth in human xenograft models. Our study highlights the rationale of targeting mitochondrial metabolism to enhance the migration and antitumor efficacy of CAR T cells in treating solid tumors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

