The pancancer overexpressed NFYC Antisense 1 controls cell cycle mitotic progression through in cis and in trans modes of action
- PMID: 38467619
- PMCID: PMC10928104
- DOI: 10.1038/s41419-024-06576-y
The pancancer overexpressed NFYC Antisense 1 controls cell cycle mitotic progression through in cis and in trans modes of action
Abstract
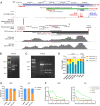
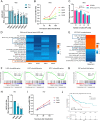
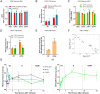
Antisense RNAs (asRNAs) represent an underappreciated yet crucial layer of gene expression regulation. Generally thought to modulate their sense genes in cis through sequence complementarity or their act of transcription, asRNAs can also regulate different molecular targets in trans, in the nucleus or in the cytoplasm. Here, we performed an in-depth molecular characterization of NFYC Antisense 1 (NFYC-AS1), the asRNA transcribed head-to-head to NFYC subunit of the proliferation-associated NF-Y transcription factor. Our results show that NFYC-AS1 is a prevalently nuclear asRNA peaking early in the cell cycle. Comparative genomics suggests a narrow phylogenetic distribution, with a probable origin in the common ancestor of mammalian lineages. NFYC-AS1 is overexpressed pancancer, preferentially in association with RB1 mutations. Knockdown of NFYC-AS1 by antisense oligonucleotides impairs cell growth in lung squamous cell carcinoma and small cell lung cancer cells, a phenotype recapitulated by CRISPR/Cas9-deletion of its transcription start site. Surprisingly, expression of the sense gene is affected only when endogenous transcription of NFYC-AS1 is manipulated. This suggests that regulation of cell proliferation is at least in part independent of the in cis transcription-mediated effect on NFYC and is possibly exerted by RNA-dependent in trans effects converging on the regulation of G2/M cell cycle phase genes. Accordingly, NFYC-AS1-depleted cells are stuck in mitosis, indicating defects in mitotic progression. Overall, NFYC-AS1 emerged as a cell cycle-regulating asRNA with dual action, holding therapeutic potential in different cancer types, including the very aggressive RB1-mutated tumors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Integrating datasets to dissect NFYC-AS1 RNA- and transcription-dependent functions: comparative transcriptome profiling of knockdown strategies.Data Brief. 2025 Apr 15;60:111565. doi: 10.1016/j.dib.2025.111565. eCollection 2025 Jun. Data Brief. 2025. PMID: 40322504 Free PMC article.
-
OIP5-AS1 Promotes Proliferation of Non-small-cell Lung Cancer and Head and Neck Squamous Cell Carcinoma Cells.Cancer Genomics Proteomics. 2021 Jul-Aug;18(4):543-548. doi: 10.21873/cgp.20279. Cancer Genomics Proteomics. 2021. PMID: 34183387 Free PMC article.
-
Long noncoding antisense RNA FAM83A-AS1 promotes lung cancer cell progression by increasing FAM83A.J Cell Biochem. 2019 Jun;120(6):10505-10512. doi: 10.1002/jcb.28336. Epub 2019 Jan 18. J Cell Biochem. 2019. PMID: 30659636 Free PMC article.
-
EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer.Cancer Res. 2018 Oct 15;78(20):5877-5890. doi: 10.1158/0008-5472.CAN-18-1011. Epub 2018 Sep 5. Cancer Res. 2018. PMID: 30185552 Free PMC article.
-
A review on the role of NCK1 Antisense RNA 1 (NCK1-AS1) in diverse disorders.Pathol Res Pract. 2023 May;245:154451. doi: 10.1016/j.prp.2023.154451. Epub 2023 Apr 5. Pathol Res Pract. 2023. PMID: 37028107 Review.
Cited by
-
Construction and validation of a prognostic model of lncRNAs associated with RNA methylation in lung adenocarcinoma.Transl Cancer Res. 2025 Feb 28;14(2):761-777. doi: 10.21037/tcr-24-1085. Epub 2025 Feb 24. Transl Cancer Res. 2025. PMID: 40104741 Free PMC article.
-
SLC5A3 depletion promotes apoptosis by inducing mitochondrial dysfunction and mitophagy in gemcitabine-resistant pancreatic cancer cells.Cell Death Dis. 2025 Mar 7;16(1):161. doi: 10.1038/s41419-025-07476-5. Cell Death Dis. 2025. PMID: 40055335 Free PMC article.
-
Integrating datasets to dissect NFYC-AS1 RNA- and transcription-dependent functions: comparative transcriptome profiling of knockdown strategies.Data Brief. 2025 Apr 15;60:111565. doi: 10.1016/j.dib.2025.111565. eCollection 2025 Jun. Data Brief. 2025. PMID: 40322504 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

