Identification of gene regulatory networks affected across drug-resistant epilepsies
- PMID: 38467626
- PMCID: PMC10928184
- DOI: 10.1038/s41467-024-46592-2
Identification of gene regulatory networks affected across drug-resistant epilepsies
Abstract
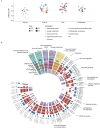
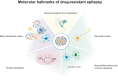
Epilepsy is a chronic and heterogenous disease characterized by recurrent unprovoked seizures, that are commonly resistant to antiseizure medications. This study applies a transcriptome network-based approach across epilepsies aiming to improve understanding of molecular disease pathobiology, recognize affected biological mechanisms and apply causal reasoning to identify therapeutic hypotheses. This study included the most common drug-resistant epilepsies (DREs), such as temporal lobe epilepsy with hippocampal sclerosis (TLE-HS), and mTOR pathway-related malformations of cortical development (mTORopathies). This systematic comparison characterized the global molecular signature of epilepsies, elucidating the key underlying mechanisms of disease pathology including neurotransmission and synaptic plasticity, brain extracellular matrix and energy metabolism. In addition, specific dysregulations in neuroinflammation and oligodendrocyte function were observed in TLE-HS and mTORopathies, respectively. The aforementioned mechanisms are proposed as molecular hallmarks of DRE with the identified upstream regulators offering opportunities for drug-target discovery and development.
© 2024. The Author(s).
Conflict of interest statement
A.R., E.A., and J.D.M. received an unrestricted grant from UCB Pharma. L.F., P.G., A.S., M.R., J.v. E., and S.D. are employees of UCB Pharma, and P.G., J.v. E., A.S., and S.D. receive stock or stock options from their employment. The remaining authors declare no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

