ST14 interacts with TMEFF1 and is a predictor of poor prognosis in ovarian cancer
- PMID: 38468232
- PMCID: PMC10929089
- DOI: 10.1186/s12885-024-11958-8
ST14 interacts with TMEFF1 and is a predictor of poor prognosis in ovarian cancer
Abstract
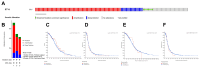
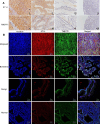
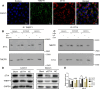
TMEFF1 is a new protein involved in the physiological functions of the central nervous system, and we previously reported TMEFF1 can promote ovarian cancer. ST14 was determined to be involved in the processes of epidermal differentiation, epithelial cell integrity, and vascular endothelial cell migration, etc. The relationship between ST14 and TMEFF1 in the ovary remains unknown. In this study, we detected the expression of ST14 and TMEFF1 in 130 different ovarian cancer tissues through immunohistochemistry. We determined ST14 and TMEFF1 were highly expressed in ovarian cancer, indicating a higher degree of tumor malignancy and a worse prognosis. Tissues significantly expressing ST14 also highly expressed TMEFF1, and the expression of the two proteins was positively correlated. Consistently, immunofluorescence double staining demonstrated the co-localization of ST14 and TMEFF1 in the same region, and immunoprecipitation confirmed the interaction between ST14 and TMEFF1. TMEFF1 expression was also reduced after knocking down ST14 through Western blot. MTT, wound healing and Transwell assays results determined that knockdown of ST14 inhibited proliferation, migration and invasion of ovarian cancer cells in vitro, but the inhibitory effect was restored after adding TMEFF1 exogenous protein. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways analysis showed that ST14 and its related genes were enriched in the processes of epithelial formation, intercellular adhesion, protein localization, and mitosis regulation. We also clarified the kinase, microRNA, and transcription factor target networks and the impact of genetic mutations on prognosis. Overall, high expression of ST14 and TMEFF1 in ovarian cancer predicts higher tumor malignancy and a worse prognosis. ST14 and TMEFF1 co-localize and interact with each other in ovarian cancer. ST14 can regulate TMEFF1 expression to promote proliferation, migration and invasion of ovarian cancer cells. We speculate that the relationship between ST14 and TMEFF1 in ovarian cancer could become a potential target for anti-cancer therapy.
Keywords: Ovarian cancer; Prognostic indicator; Protein interactions; ST14; TMEFF1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Eib DW, Holling TM, Zwijsen A, Dewulf N, de Groot E, van den Eijnden-van Raaij AJ, Huylebroeck D, Martens GJ. Expression of the follistatin/EGF-containing transmembrane protein M7365 (tomoregulin-1) during mouse development. Mech Dev. 2000;97(1–2):167–71. doi: 10.1016/S0925-4773(00)00426-3. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

