Transcriptome-wide gene expression outlier analysis pinpoints therapeutic vulnerabilities in colorectal cancer
- PMID: 38468448
- PMCID: PMC11161737
- DOI: 10.1002/1878-0261.13622
Transcriptome-wide gene expression outlier analysis pinpoints therapeutic vulnerabilities in colorectal cancer
Abstract
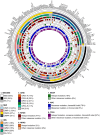
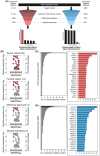
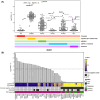
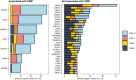
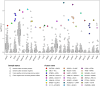
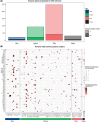
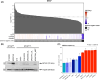
Multiple strategies are continuously being explored to expand the drug target repertoire in solid tumors. We devised a novel computational workflow for transcriptome-wide gene expression outlier analysis that allows the systematic identification of both overexpression and underexpression events in cancer cells. Here, it was applied to expression values obtained through RNA sequencing in 226 colorectal cancer (CRC) cell lines that were also characterized by whole-exome sequencing and microarray-based DNA methylation profiling. We found cell models displaying an abnormally high or low expression level for 3533 and 965 genes, respectively. Gene expression abnormalities that have been previously associated with clinically relevant features of CRC cell lines were confirmed. Moreover, by integrating multi-omics data, we identified both genetic and epigenetic alternations underlying outlier expression values. Importantly, our atlas of CRC gene expression outliers can guide the discovery of novel drug targets and biomarkers. As a proof of concept, we found that CRC cell lines lacking expression of the MTAP gene are sensitive to treatment with a PRMT5-MTA inhibitor (MRTX1719). Finally, other tumor types may also benefit from this approach.
Keywords: biomarkers; colorectal cancer; drug targets; gene expression outliers.
© 2024 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
AB served in a consulting/advisory role for Inivata and Guardant Health. AB is a member of the scientific advisory board of Neophore, Inivata, and Roche Genentech CRC Advisory Board. AB receipts grants/research supports from Neophore, Astrazeneca, and Boehringer. AB is cofounder and shareholder of NeoPhore Limited and shareholder of Kither. FDN received speaker's fees from Illumina and served in a consulting/advisory role for Amgen and Pierre Fabre Pharma. SA reports personal fees from MSD Italia and a patent (international PCT patent application No. WO 2023/199255 and Italian patent application No. 102022000007535) outside the submitted work. The remaining authors declare that they have no competing interests.
Figures

References
-
- Andrei P, Battuello P, Grasso G, Rovera E, Tesio N, Bardelli A. Integrated approaches for precision oncology in colorectal cancer: the more you know, the better. Semin Cancer Biol. 2022;84:199–213. - PubMed
-
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. - PubMed
-
- Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

