Genome-wide association study of idiopathic hypersomnia in a Japanese population
- PMID: 38469065
- PMCID: PMC10899960
- DOI: 10.1007/s41105-021-00349-2
Genome-wide association study of idiopathic hypersomnia in a Japanese population
Abstract
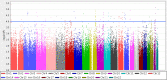
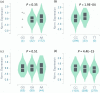
Idiopathic hypersomnia (IH) is a rare sleep disorder characterized by excessive daytime sleepiness, great difficulty upon awakening, and prolonged sleep time. In contrast to narcolepsy type 1, which is a well-recognized hypersomnia, the etiology of IH remains poorly understood. No susceptibility loci for IH have been identified, although familial aggregations have been observed among patients with IH. Narcolepsy type 1 is strongly associated with human leukocyte antigen (HLA)-DQB1*06:02; however, no significant associations between IH and HLA alleles have been reported. To identify genetic variants that affect susceptibility to IH, we performed a genome-wide association study (GWAS) and two replication studies involving a total of 414 Japanese patients with IH and 6587 healthy Japanese individuals. A meta-analysis of the three studies found no single-nucleotide polymorphisms (SNPs) that reached the genome-wide significance level. However, we identified several candidate SNPs for IH. For instance, a common genetic variant (rs2250870) within an intron of PDE9A was suggestively associated with IH. rs2250870 was significantly associated with expression levels of PDE9A in not only whole blood but also brain tissues. The leading SNP in the PDE9A region was the same in associations with both IH and PDE9A expression. PDE9A is a potential target in the treatment of several brain diseases, such as depression, schizophrenia, and Alzheimer's disease. It will be necessary to examine whether PDE9A inhibitors that have demonstrated effects on neurophysiologic and cognitive function can contribute to the development of new treatments for IH, as higher expression levels of PDE9A were observed with regard to the risk allele of rs2250870. The present study constitutes the first GWAS of genetic variants associated with IH. A larger replication study will be required to confirm these associations.
Supplementary information: The online version contains supplementary material available at 10.1007/s41105-021-00349-2.
Keywords: Genome-wide association study (GWAS); Idiopathic hypersomnia; Phosphodiesterase; Single-nucleotide polymorphisms (SNPs); Sleep.
© The Author(s), under exclusive licence to Japanese Society of Sleep Research 2021.
Conflict of interest statement
Conflict of interestDr. Inoue Yuichi has received grants and payment for lectures, including service on speakers’ bureaus, and has provided expert testimony for MSD K.K., Takeda Pharmaceutical Co. Ltd., and Eisai Co. Ltd. Dr. Makoto Honda has received consulting fees from Takeda Pharmaceutical Co. Ltd. The other authors have no competing interests to declare.
Figures

References
-
- Billiard M, Sonka K. Idiopathic hypersomnia. Sleep Med Rev. 2016;29:23–33. - PubMed
-
- Bassetti C, Aldrich MS. Idiopathic hypersomnia. A series of 42 patients. Brain. 1997;120(Pt 8):1423–1435. - PubMed
-
- Arnulf I, Leu-Semenescu S, Dodet P. Precision medicine for idiopathic hypersomnia. Sleep Med Clin. 2019;14:333–350. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
