This is a preprint.
The PCM scaffold enables RNA localization to centrosomes
- PMID: 38469150
- PMCID: PMC10926663
- DOI: 10.1101/2024.01.13.575509
The PCM scaffold enables RNA localization to centrosomes
Update in
-
The PCM scaffold enables RNA localization to centrosomes.Mol Biol Cell. 2025 Jun 1;36(6):ar75. doi: 10.1091/mbc.E25-03-0117. Epub 2025 Apr 30. Mol Biol Cell. 2025. PMID: 40305119
Abstract
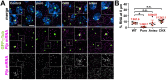
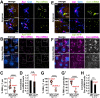
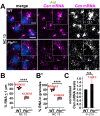
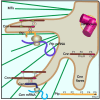
As microtubule-organizing centers, centrosomes direct assembly of the bipolar mitotic spindle required for chromosome segregation and genome stability. Centrosome activity requires the dynamic assembly of pericentriolar material (PCM), the composition and organization of which changes throughout the cell cycle. Recent studies highlight the conserved localization of several mRNAs encoded from centrosome-associated genes enriched at centrosomes, including Pericentrin-like protein (Plp) mRNA. However, relatively little is known about how RNAs localize to centrosomes and influence centrosome function. Here, we examine mechanisms underlying the subcellular localization of Plp mRNA. We find that Plp mRNA localization is puromycin-sensitive, and the Plp coding sequence is both necessary and sufficient for RNA localization, consistent with a co-translational transport mechanism. We identify regions within the Plp coding sequence that regulate Plp mRNA localization. Finally, we show that protein-protein interactions critical for elaboration of the PCM scaffold permit RNA localization to centrosomes. Taken together, these findings inform the mechanistic basis of Plp mRNA localization and lend insight into the oscillatory enrichment of RNA at centrosomes.
Keywords: Pericentrin; RNA localization; centrosome; co-translational transport.
Conflict of interest statement
Competing interest statement The authors have no competing interests to declare.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
