The Mla system and its role in maintaining outer membrane barrier function in Stenotrophomonas maltophilia
- PMID: 38469346
- PMCID: PMC10925693
- DOI: 10.3389/fcimb.2024.1346565
The Mla system and its role in maintaining outer membrane barrier function in Stenotrophomonas maltophilia
Abstract
Stenotrophomonas maltophilia are ubiquitous Gram-negative bacteria found in both natural and clinical environments. It is a remarkably adaptable species capable of thriving in various environments, thanks to the plasticity of its genome and a diverse array of genes that encode a wide range of functions. Among these functions, one notable trait is its remarkable ability to resist various antimicrobial agents, primarily through mechanisms that regulate the diffusion across cell membranes. We have investigated the Mla ABC transport system of S. maltophilia, which in other Gram-negative bacteria is known to transport phospholipids across the periplasm and is involved in maintaining outer membrane homeostasis. First, we structurally and functionally characterized the periplasmic substrate-binding protein MlaC, which determines the specificity of this system. The predicted structure of the S. maltophilia MlaC protein revealed a hydrophobic cavity of sufficient size to accommodate the phospholipids commonly found in this species. Moreover, recombinant MlaC produced heterologously demonstrated the ability to bind phospholipids. Gene knockout experiments in S. maltophilia K279a revealed that the Mla system is involved in baseline resistance to antimicrobial and antibiofilm agents, especially those with divalent-cation chelating activity. Co-culture experiments with Pseudomonas aeruginosa also showed a significant contribution of this system to the cooperation between both species in the formation of polymicrobial biofilms. As suggested for other Gram-negative pathogenic microorganisms, this system emerges as an appealing target for potential combined antimicrobial therapies.
Keywords: Mla system; Stenotrophomonas maltophilia; biofilm; chelating agents; membrane permeability.
Copyright © 2024 Coves, Mamat, Conchillo-Solé, Huedo, Bravo, Gómez, Krohn, Streit, Schaible, Gibert, Daura and Yero.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

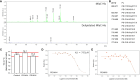
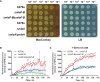

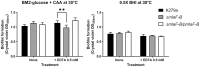

References
-
- Alcaraz E., García C., Friedman L., de Rossi B. P. (2019). The rpf/DSF signalling system of Stenotrophomonas maltophilia positively regulates biofilm formation, production of virulence-associated factors and β-lactamase induction. FEMS Microbiol. Lett. 366, fnz069. doi: 10.1093/femsle/fnz069 - DOI - PubMed
-
- Alio I., Gudzuhn M., Pérez García P., Danso D., Schoelmerich M. C., Mamat U., et al. (2020). Phenotypic and transcriptomic analyses of seven clinical Stenotrophomonas maltophilia isolates identify a small set of shared and commonly regulated genes involved in the biofilm lifestyle. Appl. Environ. Microbiol. 86, e02038–e02020. doi: 10.1128/AEM.02038-20 - DOI - PMC - PubMed
-
- Alio I., Moll R., Hoffmann T., Mamat U., Schaible U. E., Pappenfort K., et al. (2023). Stenotrophomonas maltophilia affects the gene expression profiles of the major pathogens Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro multispecies biofilm model. Microbiol. Spectr. 11, e0085923. doi: 10.1128/spectrum.00859-23 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

