Midgut serine proteinases participate in dietary adaptations of the castor (Eri) silkworm Samia ricini Anderson transferred from Ricinus communis to an ancestral host, Ailanthus excelsa Roxb
- PMID: 38469493
- PMCID: PMC10926435
- DOI: 10.3389/finsc.2023.1169596
Midgut serine proteinases participate in dietary adaptations of the castor (Eri) silkworm Samia ricini Anderson transferred from Ricinus communis to an ancestral host, Ailanthus excelsa Roxb
Abstract
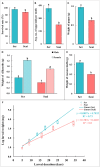
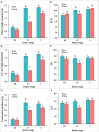
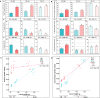
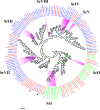
Dietary change influenced the life-history traits, nutritional utilization, and midgut serine proteinases in the larvae of the domesticated polyphagous S. ricini, transferred from R. communis (common name: castor; family Euphorbiaceae; the host plant implicated in its domestication) to A. excelsa (common name: Indian tree of heaven; family Simaroubaceae; an ancestral host of wild Samia species). Significantly higher values for fecundity and body weight were observed in larvae feeding on R. communis (Scr diet), and they took less time to reach pupation than insects feeding on A. excelsa (Scai diet). Nevertheless, the nutritional index for efficiency of conversion of digested matter (ECD) was similar for larvae feeding on the two plant species, suggesting the physiological adaptation of S. ricini (especially older instars) to an A. excelsa diet. In vitro protease assays and gelatinolytic zymograms using diagnostic substrates and protease inhibitors revealed significantly elevated levels (p ≤ 0.05) of digestive trypsins, which may be associated with the metabolic costs influencing slow growth in larvae feeding on A. excelsa. RT-PCR with semidegenerate serine proteinase gene-specific primers, and cloning and sequencing of 3' cDNA ends identified a large gene family comprising at least two groups of putative chymotrypsins (i.e., Sr I and Sr II) resembling invertebrate brachyurins/collagenases with wide substrate specificities, and five groups of putative trypsins (i.e., Sr III, Sr IV, Sr V, Sr VII, and Sr VIII). Quantitative RT-PCR indicated that transcripts belonging to the Sr I, Sr III, Sr IV, and Sr V groups, especially the Sr IV group (resembling achelase I from Lonomia achelous), were expressed differentially in the midguts of fourth instars reared on the two plant species. Sequence similarity indicated shared lineages with lepidopteran orthologs associated with expression in the gut, protein digestion, and phytophagy. The results obtained are discussed in the context of larval serine proteinases in dietary adaptations, domestication, and exploration of new host plant species for commercial rearing of S. ricini.
Keywords: digestive physiology; domestication; host plant choice; larval gut gene expression; non-mulberry silkworm; nutrition; performance; serine proteinases.
Copyright © 2023 Kashung, Bhardwaj, Saikia and Mazumdar-Leighton.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Consequences of 'no-choice, fixed time' reciprocal host plant switches on nutrition and gut serine protease gene expression in Pieris brassicae L. (Lepidoptera: Pieridae).PLoS One. 2021 Jan 20;16(1):e0245649. doi: 10.1371/journal.pone.0245649. eCollection 2021. PLoS One. 2021. PMID: 33471847 Free PMC article.
-
Expression of diverse midgut serine proteinases in the sericigenous Lepidoptera Antheraea assamensis (Helfer) is influenced by choice of host plant species.Insect Mol Biol. 2011 Feb;20(1):1-13. doi: 10.1111/j.1365-2583.2010.01048.x. Epub 2010 Sep 21. Insect Mol Biol. 2011. PMID: 20854480
-
Host hemolymph proteins and protein digestion in larval Habrobracon hebetor (Hymenoptera: braconidae).Insect Biochem Mol Biol. 2000 Oct;30(10):937-46. doi: 10.1016/s0965-1748(00)00066-7. Insect Biochem Mol Biol. 2000. PMID: 10899460
-
Differential effects of sugar-mimic alkaloids in mulberry latex on sugar metabolism and disaccharidases of Eri and domesticated silkworms: enzymatic adaptation of Bombyx mori to mulberry defense.Insect Biochem Mol Biol. 2007 Dec;37(12):1348-58. doi: 10.1016/j.ibmb.2007.09.001. Epub 2007 Sep 12. Insect Biochem Mol Biol. 2007. PMID: 17967353
-
Identification of six chymotrypsin cDNAs from larval midguts of Helicoverpa zea and Agrotis ipsilon feeding on the soybean (Kunitz) trypsin inhibitor.Insect Biochem Mol Biol. 2001 Apr 27;31(6-7):633-44. doi: 10.1016/s0965-1748(00)00168-5. Insect Biochem Mol Biol. 2001. PMID: 11267902
References
-
- Peigler RS, Naumann S. A revision of the silkmoth genus Samia. San Antonio, TX: University of the Incarnate Word; (2003) p. 55–70.
-
- Piegler RS, Calhoun JV. Correct authorship of the name Phalaena ricini and the nomenclatural status of the name Saturnia canningi (Lepidoptera: Saturniidae). Trop Lepid Res (2013) 23:39–43.
-
- Roxburgh W, III. Account of the tusseh and arrindy silk-worms of Bengal. Trans Linn Soc London (1804) 7:33–48. doi: 10.1111/j.1096-3642.1804.tb00278.x - DOI
-
- Chowdhury SN. Biology of silkworm and host plants Vol. 28. . Dibrugarh, Assam: GB Publishers; (2005).
LinkOut - more resources
Full Text Sources
Research Materials

