Transcriptomic signatures of progressive and regressive liver fibrosis and portal hypertension
- PMID: 38469563
- PMCID: PMC10926212
- DOI: 10.1016/j.isci.2024.109301
Transcriptomic signatures of progressive and regressive liver fibrosis and portal hypertension
Abstract
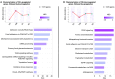
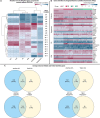
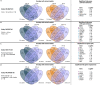
Persistent liver injury triggers a fibrogenic program that causes pathologic remodeling of the hepatic microenvironment (i.e., liver fibrosis) and portal hypertension. The dynamics of gene regulation during liver disease progression and early regression remain understudied. Here, we generated hepatic transcriptome profiles in two well-established liver disease models at peak fibrosis and during spontaneous regression after the removal of the inducing agents. We linked the dynamics of key disease readouts, such as portal pressure, collagen area, and transaminase levels, to differentially expressed genes, enabling the identification of transcriptomic signatures of progressive vs. regressive liver fibrosis and portal hypertension. These candidate biomarkers (e.g., Tcf4, Mmp7, Trem2, Spp1, Scube1, Islr) were validated in RNA sequencing datasets of patients with cirrhosis and portal hypertension, and those cured from hepatitis C infection. Finally, deconvolution identified major cell types and suggested an association of macrophage and portal hepatocyte signatures with portal hypertension and fibrosis area.
Keywords: Disease; Fibrosis; Integrative aspects of cell biology; Model organism; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T., Portman J.R., Matchett K.P., Brice M., Marwick J.A., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. - DOI - PMC - PubMed
-
- Thanapirom K., Suksawatamnuay S., Tanpowpong N., Chaopathomkul B., Sriphoosanaphan S., Thaimai P., Srisoonthorn N., Treeprasertsuk S., Komolmit P. Non-invasive tests for liver fibrosis assessment in patients with chronic liver diseases: a prospective study. Sci. Rep. 2022;12:4913. doi: 10.1038/s41598-022-08955-x. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

