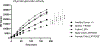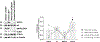Biallelic USP14 variants cause a syndromic neurodevelopmental disorder
- PMID: 38469793
- PMCID: PMC11241549
- DOI: 10.1016/j.gim.2024.101120
Biallelic USP14 variants cause a syndromic neurodevelopmental disorder
Abstract
Purpose: Imbalances in protein homeostasis affect human brain development, with the ubiquitin-proteasome system (UPS) and autophagy playing crucial roles in neurodevelopmental disorders (NDD). This study explores the impact of biallelic USP14 variants on neurodevelopment, focusing on its role as a key hub connecting UPS and autophagy.
Methods: Here, we identified biallelic USP14 variants in 4 individuals from 3 unrelated families: 1 fetus, a newborn with a syndromic NDD and 2 siblings affected by a progressive neurological disease. Specifically, the 2 siblings from the latter family carried 2 compound heterozygous variants c.8T>C p.(Leu3Pro) and c.988C>T p.(Arg330∗), whereas the fetus had a homozygous frameshift c.899_902del p.(Lys300Serfs∗24) variant, and the newborn patient harbored a homozygous frameshift c.233_236del p.(Leu78Glnfs∗11) variant. Functional studies were conducted using sodium dodecyl-sulfate polyacrylamide gel electrophoresis, western blotting, and mass spectrometry analyses in both patient-derived and CRISPR-Cas9-generated cells.
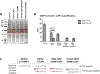
Results: Our investigations indicated that the USP14 variants correlated with reduced N-terminal methionine excision, along with profound alterations in proteasome, autophagy, and mitophagy activities.
Conclusion: Biallelic USP14 variants in NDD patients perturbed protein degradation pathways, potentially contributing to disorder etiology. Altered UPS, autophagy, and mitophagy activities underscore the intricate interplay, elucidating their significance in maintaining proper protein homeostasis during brain development.
Keywords: Loss-of-function variants; N-terminal methionine excision; Neurodevelopmental disorders; USP14; Ubiquitin-proteasome system.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures