Formation of a constructed microbial community in a nutrient-rich environment indicates bacterial interspecific competition
- PMID: 38470038
- PMCID: PMC11019790
- DOI: 10.1128/msystems.00006-24
Formation of a constructed microbial community in a nutrient-rich environment indicates bacterial interspecific competition
Abstract
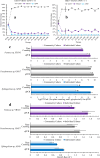
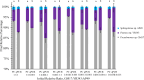
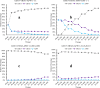
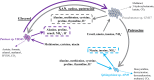
Understanding the organizational principles of microbial communities is essential for interpreting ecosystem stability. Previous studies have investigated the formation of bacterial communities under nutrient-poor conditions or obligate relationships to observe cooperative interactions among different species. How microorganisms form stabilized communities in nutrient-rich environments, without obligate metabolic interdependency for growth, is still not fully disclosed. In this study, three bacterial strains isolated from the Populus deltoides rhizosphere were co-cultured in complex medium, and their growth behavior was tracked. These strains co-exist in mixed culture over serial transfer for multiple growth-dilution cycles. Competition is proposed as an emergent interaction relationship among the three bacteria based on their significantly decreased growth levels. The effects of different initial inoculum ratios, up to three orders of magnitude, on community structure were investigated, and the final compositions of the mixed communities with various starting composition indicate that community structure is not dependent on the initial inoculum ratio. Furthermore, the competitive relationships within the community were not altered by different initial inoculum ratios. The community structure was simulated by generalized Lotka-Volterra and dynamic flux balance analysis to provide mechanistic predictions into emergence of community structure under a nutrient-rich environment. Metaproteomic analyses provide support for the metabolite exchanges predicted by computational modeling and for highly altered physiologies when microbes are grown in co-culture. These findings broaden our understanding of bacterial community dynamics and metabolic diversity in higher-order interactions and could be significant in the management of rhizospheric bacterial communities.
Importance: Bacteria naturally co-exist in multispecies consortia, and the ability to engineer such systems can be useful in biotechnology. Despite this, few studies have been performed to understand how bacteria form a stable community and interact with each other under nutrient-rich conditions. In this study, we investigated the effects of initial inoculum ratios on bacterial community structure using a complex medium and found that the initial inoculum ratio has no significant impact on resultant community structure or on interaction patterns between community members. The microbial population profiles were simulated using computational tools in order to understand intermicrobial relationships and to identify potential metabolic exchanges that occur during stabilization of the bacterial community. Studying microbial community assembly processes is essential for understanding fundamental ecological principles in microbial ecosystems and can be critical in predicting microbial community structure and function.
Keywords: dynamic flux balance analysis; initial inoculum ratio; metabolite exchange; metaproteomics; microbial community; rhizospheric bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Metaproteomics reveals insights into microbial structure, interactions, and dynamic regulation in defined communities as they respond to environmental disturbance.BMC Microbiol. 2021 Nov 8;21(1):308. doi: 10.1186/s12866-021-02370-4. BMC Microbiol. 2021. PMID: 34749649 Free PMC article.
-
Formation, characterization and modeling of emergent synthetic microbial communities.Comput Struct Biotechnol J. 2021 Apr 9;19:1917-1927. doi: 10.1016/j.csbj.2021.03.034. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33995895 Free PMC article.
-
Nutrient levels and trade-offs control diversity in a serial dilution ecosystem.Elife. 2020 Sep 11;9:e57790. doi: 10.7554/eLife.57790. Elife. 2020. PMID: 32915132 Free PMC article.
-
Metabolic ecology of microbiomes: Nutrient competition, host benefits, and community engineering.Cell Host Microbe. 2025 Jun 11;33(6):790-807. doi: 10.1016/j.chom.2025.05.013. Cell Host Microbe. 2025. PMID: 40505619 Review.
-
Rhizospheric microbiome: Bio-based emerging strategies for sustainable agriculture development and future perspectives.Microbiol Res. 2022 Jan;254:126901. doi: 10.1016/j.micres.2021.126901. Epub 2021 Oct 23. Microbiol Res. 2022. PMID: 34700186 Review.
Cited by
-
The impact of altered dietary adenine concentrations on the gut microbiota in Drosophila.Front Microbiol. 2024 Aug 5;15:1433155. doi: 10.3389/fmicb.2024.1433155. eCollection 2024. Front Microbiol. 2024. PMID: 39161604 Free PMC article.
-
Sustainable Food Waste Management in Anaerobic Digesters: Prediction of the Organic Load Impact by Metagenome-Scale Metabolic Modeling.Environ Sci Technol. 2025 Apr 8;59(13):6659-6672. doi: 10.1021/acs.est.4c11180. Epub 2025 Mar 24. Environ Sci Technol. 2025. PMID: 40126624 Free PMC article.
-
Quorum sensing modulates microbial community structure through regulation of secondary metabolites.mSphere. 2025 Jul 29;10(7):e0105024. doi: 10.1128/msphere.01050-24. Epub 2025 Jun 20. mSphere. 2025. PMID: 40539822 Free PMC article.
References
-
- Prashar P, Kapoor N, Sachdeva S. 2014. Rhizosphere: its structure, bacterial diversity and significance. Rev Environ Sci Biotechnol 13:63–77. doi:10.1007/s11157-013-9317-z - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
