The recruitment of TRiC chaperonin in rotavirus viroplasms correlates with virus replication
- PMID: 38470055
- PMCID: PMC11005421
- DOI: 10.1128/mbio.00499-24
The recruitment of TRiC chaperonin in rotavirus viroplasms correlates with virus replication
Abstract
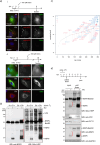
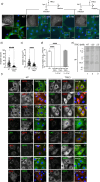
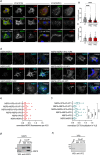
Rotavirus (RV) replication takes place in the viroplasms, cytosolic inclusions that allow the synthesis of virus genome segments and their encapsidation in the core shell, followed by the addition of the second layer of the virion. The viroplasms are composed of several viral proteins, including NSP5, which serves as the main building block. Microtubules, lipid droplets, and miRNA-7 are among the host components recruited in viroplasms. We investigated the interaction between RV proteins and host components of the viroplasms by performing a pull-down assay of lysates from RV-infected cells expressing NSP5-BiolD2. Subsequent tandem mass spectrometry identified all eight subunits of the tailless complex polypeptide I ring complex (TRiC), a cellular chaperonin responsible for folding at least 10% of the cytosolic proteins. Our confirmed findings reveal that TRiC is brought into viroplasms and wraps around newly formed double-layered particles. Chemical inhibition of TRiC and silencing of its subunits drastically reduced virus progeny production. Through direct RNA sequencing, we show that TRiC is critical for RV replication by controlling dsRNA genome segment synthesis, particularly negative-sense single-stranded RNA. Importantly, cryo-electron microscopy analysis shows that TRiC inhibition results in defective virus particles lacking genome segments and polymerase complex (VP1/VP3). Moreover, TRiC associates with VP2 and NSP5 but not with VP1. Also, VP2 is shown to be essential for recruiting TRiC in viroplasms and preserving their globular morphology. This study highlights the essential role of TRiC in viroplasm formation and in facilitating virion assembly during the RV life cycle.
Importance: The replication of rotavirus takes place in cytosolic inclusions termed viroplasms. In these inclusions, the distinct 11 double-stranded RNA genome segments are co-packaged to complete a genome in newly generated virus particles. In this study, we show for the first time that the tailless complex polypeptide I ring complex (TRiC), a cellular chaperonin responsible for the folding of at least 10% of the cytosolic proteins, is a component of viroplasms and is required for the synthesis of the viral negative-sense single-stranded RNA. Specifically, TRiC associates with NSP5 and VP2, the cofactor involved in RNA replication. Our study adds a new component to the current model of rotavirus replication, where TRiC is recruited to viroplasms to assist replication.
Keywords: NSP5; TRiC; VP2; chaperones; double-stranded RNA virus; rotavirus; viral replication; viroplasm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
