Dysregulated proteome and N-glycoproteome in ALG1-deficient fibroblasts
- PMID: 38470198
- PMCID: PMC7616334
- DOI: 10.1002/pmic.202400012
Dysregulated proteome and N-glycoproteome in ALG1-deficient fibroblasts
Abstract
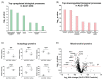
Asparagine-linked glycosylation 1 protein is a β-1,4-mannosyltransferase, is encoded by the ALG1 gene, which catalyzes the first step of mannosylation in N-glycosylation. Pathogenic variants in ALG1 cause a rare autosomal recessive disorder termed as ALG1-CDG. We performed a quantitative proteomics and N-glycoproteomics study in fibroblasts derived from patients with one homozygous and two compound heterozygous pathogenic variants in ALG1. Several proteins that exhibited significant upregulation included insulin-like growth factor II and pleckstrin, whereas hyaluronan and proteoglycan link protein 1 was downregulated. These proteins are crucial for cell growth, survival and differentiation. Additionally, we observed a decrease in the expression of mitochondrial proteins and an increase in autophagy-related proteins, suggesting mitochondrial and cellular stress. N-glycoproteomics revealed the reduction in high-mannose and complex/hybrid glycopeptides derived from numerous proteins in patients explaining that defect in ALG1 has broad effects on glycosylation. Further, we detected an increase in several short oligosaccharides, including chitobiose (HexNAc2) trisaccharides (Hex-HexNAc2) and novel tetrasaccharides (NeuAc-Hex-HexNAc2) derived from essential proteins including LAMP1, CD44 and integrin. These changes in glycosylation were observed in all patients irrespective of their gene variants. Overall, our findings not only provide novel molecular insights into understanding ALG1-CDG but also offer short oligosaccharide-bearing peptides as potential biomarkers.
Keywords: ALG1‐CDG; LC‐MS/MS; biomarker; congenital disorders of glycosylation; glycoproteomics; proteomics.
© 2024 The Authors. PROTEOMICS published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
ALG1-CDG: Clinical and Molecular Characterization of 39 Unreported Patients.Hum Mutat. 2016 Jul;37(7):653-60. doi: 10.1002/humu.22983. Epub 2016 Mar 21. Hum Mutat. 2016. PMID: 26931382 Free PMC article.
-
A Novel N-Tetrasaccharide in Patients with Congenital Disorders of Glycosylation, Including Asparagine-Linked Glycosylation Protein 1, Phosphomannomutase 2, and Mannose Phosphate Isomerase Deficiencies.Clin Chem. 2016 Jan;62(1):208-17. doi: 10.1373/clinchem.2015.243279. Epub 2015 Oct 1. Clin Chem. 2016. PMID: 26430078 Free PMC article.
-
N-glycoproteomic and proteomic alterations in SRD5A3-deficient fibroblasts.Glycobiology. 2024 Sep 30;34(11):cwae076. doi: 10.1093/glycob/cwae076. Glycobiology. 2024. PMID: 39360848 Free PMC article.
-
A novel variant in ALG1 gene associated with congenital disorder of glycosylation: A case report and short literature review.Mol Genet Genomic Med. 2023 Aug;11(8):e2197. doi: 10.1002/mgg3.2197. Epub 2023 May 19. Mol Genet Genomic Med. 2023. PMID: 37204045 Free PMC article. Review.
-
Novel variants and clinical symptoms in four new ALG3-CDG patients, review of the literature, and identification of AAGRP-ALG3 as a novel ALG3 variant with alanine and glycine-rich N-terminus.Hum Mutat. 2019 Jul;40(7):938-951. doi: 10.1002/humu.23764. Epub 2019 May 8. Hum Mutat. 2019. PMID: 31067009 Review.
Cited by
-
Deciphering the Glycoproteomic Landscape of Mood Disorders: Unveiling Molecular Association Between CDG and Depression Resilience.Res Sq [Preprint]. 2025 Jul 10:rs.3.rs-6882753. doi: 10.21203/rs.3.rs-6882753/v1. Res Sq. 2025. PMID: 40671800 Free PMC article. Preprint.
-
Predicting disease-overarching therapeutic approaches for congenital disorders of glycosylation using multi-OMICS.Mol Genet Metab. 2025 Jul 19;146(1-2):109195. doi: 10.1016/j.ymgme.2025.109195. Online ahead of print. Mol Genet Metab. 2025. PMID: 40743674
-
Advancement in Clinical Glycomics and Glycoproteomics for Congenital Disorders of Glycosylation: Progress and Challenges Ahead.Biomedicines. 2025 Aug 13;13(8):1964. doi: 10.3390/biomedicines13081964. Biomedicines. 2025. PMID: 40868218 Free PMC article. Review.
-
The N-glycosylation defect in Lec5 and Lec9 CHO cells is caused by absence of the DHRSX gene.bioRxiv [Preprint]. 2024 Jun 18:2024.06.18.599300. doi: 10.1101/2024.06.18.599300. bioRxiv. 2024. Update in: J Biol Chem. 2024 Dec;300(12):107875. doi: 10.1016/j.jbc.2024.107875. PMID: 38948797 Free PMC article. Updated. Preprint.
-
Liposome-encapsulated mannose-1-phosphate therapy improves global N-glycosylation in different congenital disorders of glycosylation.Mol Genet Metab. 2024 Jun;142(2):108487. doi: 10.1016/j.ymgme.2024.108487. Epub 2024 May 7. Mol Genet Metab. 2024. PMID: 38733638 Free PMC article.
References
-
- Huffaker TC, Robbins PW. Temperature-sensitive yeast mutants deficient in asparagine-linked glycosylation. The Journal of Biological Chemistry. 1982;257:3203–3210. - PubMed
-
- Grubenmann CE. Deficiency of the first mannosylation step in the N-glycosylation pathway causes congenital disorder of glycosylation type Ik. Human Molecular Genetics. 2004;13:535–542. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

