Computational aptamer design for spike glycoprotein (S) (SARS CoV-2) detection with an electrochemical aptasensor
- PMID: 38470514
- PMCID: PMC10933206
- DOI: 10.1007/s00253-024-13066-w
Computational aptamer design for spike glycoprotein (S) (SARS CoV-2) detection with an electrochemical aptasensor
Abstract
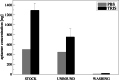
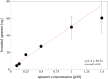
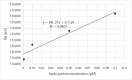
A new bioinformatic platform (APTERION) was used to design in a short time and with high specificity an aptamer for the detection of the spike protein, a structural protein of SARS-CoV-2 virus, responsible for the COVID-19 pandemic. The aptamer concentration on the carbon electrode surface was optimized using static contact angle and fluorescence method, while specificity was tested using differential pulse voltammetry (DPV) associated to carbon screen-printed electrodes. The data obtained demonstrated the good features of the aptamer which could be used to create a rapid method for the detection of SARS-CoV-2 virus. In fact, it is specific for spike also when tested against bovine serum albumin and lysozyme, competitor proteins if saliva is used as sample to test for the virus presence. Spectrofluorometric characterization allowed to measure the amount of aptamer present on the carbon electrode surface, while DPV measurements proved the affinity of the aptamer towards the spike protein and gave quantitative results. The acquired data allowed to conclude that the APTERION bioinformatic platform is a good method for aptamer design for rapidity and specificity. KEY POINTS: • Spike protein detection using an electrochemical biosensor • Aptamer characterization by contact angle and fluorescent measurements on electrode surface • Computational design of specific aptamers to speed up the aptameric sequence time.
Keywords: Aptamer; Bioinformatic platform; Biosensor; Screen-printed electrodes; Spike protein.
© 2024. The Author(s).
Conflict of interest statement
The author of this manuscript Antonello Romani is co-founder and CEO of Arta Peptidion s.r.ls and Aldo Feriani is CTO and R&D Manager of Arta Peptidion. The company had no influence on the study design or on the content of this manuscript. This commercial affiliation does not alter our adherence to policies on sharing data and materials. This work was not funded by the Arta Peptidion company.
Figures





References
-
- Asai K, Yamamoto T, Nagashima S, Ogata G, Hibino H, Einaga Y (2020) An electrochemical aptamer-based sensor prepared by utilizing the strong interaction between a DNA aptamer and diamond. Analyst 145:544–549. 10.1039/c9an01976f - PubMed
-
- Bagheryan Z, Raoof JB, Golabi M, Turner APF, Beni V (2016) Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Bios and Bioel 80:566–573. 10.1016/j.bios.2016.02.024 - PubMed
-
- Browne C, Powley EJ, Whitehouse D, Lucas S, Cowling PI, Rohlfshagen P, Tavener S, Perez Liebana D, Samothrakis S, Colton S (2012) A survey of Monte Carlo tree search methods. IEEE Trans Comput Intell AI Games 4(1):1–43. 10.1109/TCIAIG.2012.2186810
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

