A gradient of the HD-Zip regulator Woolly regulates multicellular trichome morphogenesis in tomato
- PMID: 38470570
- PMCID: PMC11132899
- DOI: 10.1093/plcell/koae077
A gradient of the HD-Zip regulator Woolly regulates multicellular trichome morphogenesis in tomato
Abstract
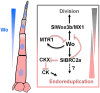
Homeodomain (HD) proteins regulate embryogenesis in animals such as the fruit fly (Drosophila melanogaster), often in a concentration-dependent manner. HD-leucine zipper (Zip) IV family genes are unique to plants and often function in the L1 epidermal cell layer. However, our understanding of the roles of HD-Zip IV family genes in plant morphogenesis is limited. In this study, we investigated the morphogenesis of tomato (Solanum lycopersicum) multicellular trichomes, a type of micro-organ in plants. We found that a gradient of the HD-Zip IV regulator Woolly (Wo) coordinates spatially polarized cell division and cell expansion in multicellular trichomes. Moreover, we identified a TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR (TCP) transcription factor-encoding gene, SlBRANCHED2a (SlBRC2a), as a key downstream target of Wo that regulates the transition from cell division to cell expansion. High levels of Wo promote cell division in apical trichome cells, whereas in basal trichome cells, Wo mediates a negative feedback loop with SlBRC2a that forces basal cells to enter endoreduplication. The restricted high and low activities of Wo pattern the morphogenesis of tomato multicellular trichomes. These findings provide insights into the functions of HD-Zip IV genes during plant morphogenesis.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement: The authors declare no competing interest.
Figures








References
-
- Anirban A. 70 years of turing patterns. Nat Rev Phys. 2022:4(7):432–432. 10.1038/s42254-022-00486-8 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

