Noncanonical and reversible cysteine ubiquitination prevents the overubiquitination of PEX5 at the peroxisomal membrane
- PMID: 38470934
- PMCID: PMC10959387
- DOI: 10.1371/journal.pbio.3002567
Noncanonical and reversible cysteine ubiquitination prevents the overubiquitination of PEX5 at the peroxisomal membrane
Abstract
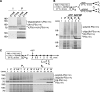
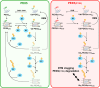
PEX5, the peroxisomal protein shuttling receptor, binds newly synthesized proteins in the cytosol and transports them to the organelle. During its stay at the peroxisomal protein translocon, PEX5 is monoubiquitinated at its cysteine 11 residue, a mandatory modification for its subsequent ATP-dependent extraction back into the cytosol. The reason why a cysteine and not a lysine residue is the ubiquitin acceptor is unknown. Using an established rat liver-based cell-free in vitro system, we found that, in contrast to wild-type PEX5, a PEX5 protein possessing a lysine at position 11 is polyubiquitinated at the peroxisomal membrane, a modification that negatively interferes with the extraction process. Wild-type PEX5 cannot retain a polyubiquitin chain because ubiquitination at cysteine 11 is a reversible reaction, with the E2-mediated deubiquitination step presenting faster kinetics than PEX5 polyubiquitination. We propose that the reversible nonconventional ubiquitination of PEX5 ensures that neither the peroxisomal protein translocon becomes obstructed with polyubiquitinated PEX5 nor is PEX5 targeted for proteasomal degradation.
Copyright: © 2024 Francisco et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway.J Biol Chem. 2009 Oct 2;284(40):27243-51. doi: 10.1074/jbc.M109.032565. Epub 2009 Jul 23. J Biol Chem. 2009. PMID: 19632994 Free PMC article.
-
Peroxisomal monoubiquitinated PEX5 interacts with the AAA ATPases PEX1 and PEX6 and is unfolded during its dislocation into the cytosol.J Biol Chem. 2018 Jul 20;293(29):11553-11563. doi: 10.1074/jbc.RA118.003669. Epub 2018 Jun 8. J Biol Chem. 2018. PMID: 29884772 Free PMC article.
-
The Extraction Mechanism of Monoubiquitinated PEX5 from the Peroxisomal Membrane.J Mol Biol. 2023 Jan 30;435(2):167896. doi: 10.1016/j.jmb.2022.167896. Epub 2022 Nov 26. J Mol Biol. 2023. PMID: 36442669
-
Ubiquitin in the peroxisomal protein import pathway.Biochimie. 2014 Mar;98:29-35. doi: 10.1016/j.biochi.2013.08.003. Epub 2013 Aug 14. Biochimie. 2014. PMID: 23954799 Review.
-
Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis.Cell Cycle. 2017;16(21):2037-2045. doi: 10.1080/15384101.2017.1376149. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933989 Free PMC article. Review.
Cited by
-
Seventy years of peroxisome research: current advances and future perspectives.Histochem Cell Biol. 2025 Jan 18;163(1):24. doi: 10.1007/s00418-024-02349-y. Histochem Cell Biol. 2025. PMID: 39825042 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

