CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23
- PMID: 38471818
- PMCID: PMC11014256
- DOI: 10.1093/nar/gkae167
CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23
Abstract
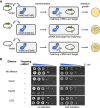
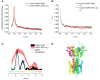
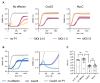
CRISPR-Cas provides adaptive immunity in prokaryotes. Type III CRISPR systems detect invading RNA and activate the catalytic Cas10 subunit, which generates a range of nucleotide second messengers to signal infection. These molecules bind and activate a diverse range of effector proteins that provide immunity by degrading viral components and/or by disturbing key aspects of cellular metabolism to slow down viral replication. Here, we focus on the uncharacterised effector Csx23, which is widespread in Vibrio cholerae. Csx23 provides immunity against plasmids and phage when expressed in Escherichia coli along with its cognate type III CRISPR system. The Csx23 protein localises in the membrane using an N-terminal transmembrane α-helical domain and has a cytoplasmic C-terminal domain that binds cyclic tetra-adenylate (cA4), activating its defence function. Structural studies reveal a tetrameric structure with a novel fold that binds cA4 specifically. Using pulse EPR, we demonstrate that cA4 binding to the cytoplasmic domain of Csx23 results in a major perturbation of the transmembrane domain, consistent with the opening of a pore and/or disruption of membrane integrity. This work reveals a new class of cyclic nucleotide binding protein and provides key mechanistic detail on a membrane-associated CRISPR effector.
Plain language summary
Many anti-viral defence systems generate a cyclic nucleotide signal that activates cellular defences in response to infection. Type III CRISPR systems use a specialised polymerase to make cyclic oligoadenylate (cOA) molecules from ATP. These can bind and activate a range of effector proteins that slow down viral replication. In this study, we focussed on the Csx23 effector from the human pathogen Vibrio cholerae – a trans-membrane protein that binds a cOA molecule, leading to anti-viral immunity. Structural studies revealed a new class of nucleotide recognition domain, where cOA binding is transmitted to changes in the trans-membrane domain, most likely resulting in membrane depolarisation. This study highlights the diversity of mechanisms for anti-viral defence via nucleotide signalling.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA4.J Biol Chem. 2020 Oct 30;295(44):14963-14972. doi: 10.1074/jbc.RA120.014099. Epub 2020 Aug 21. J Biol Chem. 2020. PMID: 32826317 Free PMC article.
-
The CRISPR effector Cam1 mediates membrane depolarization for phage defence.Nature. 2024 Jan;625(7996):797-804. doi: 10.1038/s41586-023-06902-y. Epub 2024 Jan 10. Nature. 2024. PMID: 38200316 Free PMC article.
-
Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.Elife. 2020 Jun 29;9:e57627. doi: 10.7554/eLife.57627. Elife. 2020. PMID: 32597755 Free PMC article.
-
The diverse arsenal of type III CRISPR-Cas-associated CARF and SAVED effectors.Biochem Soc Trans. 2022 Oct 31;50(5):1353-1364. doi: 10.1042/BST20220289. Biochem Soc Trans. 2022. PMID: 36282000 Free PMC article. Review.
-
Type III CRISPR-Cas: beyond the Cas10 effector complex.Trends Biochem Sci. 2024 Jan;49(1):28-37. doi: 10.1016/j.tibs.2023.10.006. Epub 2023 Nov 8. Trends Biochem Sci. 2024. PMID: 37949766 Free PMC article. Review.
Cited by
-
Structural integrity and side-chain interaction at the loop region of the bridge helix modulate Cas9 substrate discrimination.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf557. doi: 10.1093/nar/gkaf557. Nucleic Acids Res. 2025. PMID: 40613714 Free PMC article.
-
Bioinformatic analysis of type III CRISPR systems reveals key properties and new effector families.Nucleic Acids Res. 2024 Jul 8;52(12):7129-7141. doi: 10.1093/nar/gkae462. Nucleic Acids Res. 2024. PMID: 38808661 Free PMC article.
-
Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1277. doi: 10.1093/nar/gkae1277. Nucleic Acids Res. 2025. PMID: 39817514 Free PMC article.
-
SAM-AMP lyases in type III CRISPR defence.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf655. doi: 10.1093/nar/gkaf655. Nucleic Acids Res. 2025. PMID: 40650973 Free PMC article.
-
Nucleic acid recognition during prokaryotic immunity.Mol Cell. 2025 Jan 16;85(2):309-322. doi: 10.1016/j.molcel.2024.12.007. Epub 2025 Jan 16. Mol Cell. 2025. PMID: 39824170 Review.
References
-
- Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas C., Siksnys V.. Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell. 2016; 62:295–306. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

