Theileria parasites sequester host eIF5A to escape elimination by host-mediated autophagy
- PMID: 38472173
- PMCID: PMC10933305
- DOI: 10.1038/s41467-024-45022-7
Theileria parasites sequester host eIF5A to escape elimination by host-mediated autophagy
Abstract
Intracellular pathogens develop elaborate mechanisms to survive within the hostile environments of host cells. Theileria parasites infect bovine leukocytes and cause devastating diseases in cattle in developing countries. Theileria spp. have evolved sophisticated strategies to hijack host leukocytes, inducing proliferative and invasive phenotypes characteristic of cell transformation. Intracellular Theileria parasites secrete proteins into the host cell and recruit host proteins to induce oncogenic signaling for parasite survival. It is unknown how Theileria parasites evade host cell defense mechanisms, such as autophagy, to survive within host cells. Here, we show that Theileria annulata parasites sequester the host eIF5A protein to their surface to escape elimination by autophagic processes. We identified a small-molecule compound that reduces parasite load by inducing autophagic flux in host leukocytes, thereby uncoupling Theileria parasite survival from host cell survival. We took a chemical genetics approach to show that this compound induced host autophagy mechanisms and the formation of autophagic structures via AMPK activation and the release of the host protein eIF5A which is sequestered at the parasite surface. The sequestration of host eIF5A to the parasite surface offers a strategy to escape elimination by autophagic mechanisms. These results show how intracellular pathogens can avoid host defense mechanisms and identify a new anti-Theileria drug that induces autophagy to target parasite removal.
© 2024. The Author(s).
Conflict of interest statement
All authors do not have any competing interests.
Figures


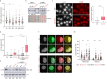
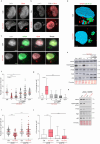
References
MeSH terms
Grants and funding
- ANR-15-CE12-0020/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-EURE-0013/Agence Nationale de la Recherche (French National Research Agency)
- ANR-11-LABX-0071/Agence Nationale de la Recherche (French National Research Agency)
- ANR-11-IDEX-0005-01/Agence Nationale de la Recherche (French National Research Agency)
- #EQU202203014701/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

