A single nuclear transcriptomic characterisation of mechanisms responsible for impaired angiogenesis and blood-brain barrier function in Alzheimer's disease
- PMID: 38472200
- PMCID: PMC10933340
- DOI: 10.1038/s41467-024-46630-z
A single nuclear transcriptomic characterisation of mechanisms responsible for impaired angiogenesis and blood-brain barrier function in Alzheimer's disease
Abstract
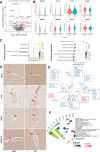
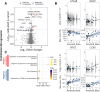
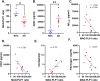
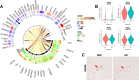
Brain perfusion and blood-brain barrier (BBB) integrity are reduced early in Alzheimer's disease (AD). We performed single nucleus RNA sequencing of vascular cells isolated from AD and non-diseased control brains to characterise pathological transcriptional signatures responsible for this. We show that endothelial cells (EC) are enriched for expression of genes associated with susceptibility to AD. Increased β-amyloid is associated with BBB impairment and a dysfunctional angiogenic response related to a failure of increased pro-angiogenic HIF1A to increased VEGFA signalling to EC. This is associated with vascular inflammatory activation, EC senescence and apoptosis. Our genomic dissection of vascular cell risk gene enrichment provides evidence for a role of EC pathology in AD and suggests that reducing vascular inflammatory activation and restoring effective angiogenesis could reduce vascular dysfunction contributing to the genesis or progression of early AD.
© 2024. The Author(s).
Conflict of interest statement
P.M.M. is a consultant for Biogen, Sudo Therapeutics, Nimbus, Astex, GSK and Sangamo. He has received research funding for aspects of this work from Biogen and the UK DRI. He has research funding unrelated to this work from Biogen and Bristol Meyers Squibb. ZC is founder and director of Oxford StemTech and Human-Centric DD. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

