Quantitative proteomics reveals tissue-specific, infection-induced and species-specific neutrophil protein signatures
- PMID: 38472281
- PMCID: PMC10933280
- DOI: 10.1038/s41598-024-56163-6
Quantitative proteomics reveals tissue-specific, infection-induced and species-specific neutrophil protein signatures
Abstract
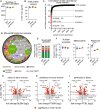
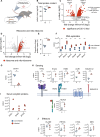
Neutrophils are one of the first responders to infection and are a key component of the innate immune system through their ability to phagocytose and kill invading pathogens, secrete antimicrobial molecules and produce extracellular traps. Neutrophils are produced in the bone marrow, circulate within the blood and upon immune challenge migrate to the site of infection. We wanted to understand whether this transition shapes the mouse neutrophil protein landscape, how the mouse neutrophil proteome is impacted by systemic infection and perform a comparative analysis of human and mouse neutrophils. Using quantitative mass spectrometry we reveal tissue-specific, infection-induced and species-specific neutrophil protein signatures. We show a high degree of proteomic conservation between mouse bone marrow, blood and peritoneal neutrophils, but also identify key differences in the molecules that these cells express for sensing and responding to their environment. Systemic infection triggers a change in the bone marrow neutrophil population with considerable impact on the core machinery for protein synthesis and DNA replication along with environmental sensors. We also reveal profound differences in mouse and human blood neutrophils, particularly their granule contents. Our proteomics data provides a valuable resource for understanding neutrophil function and phenotypes across species and model systems.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

