A pan-cancer analysis of the oncogenic and immunological roles of transglutaminase 1 (TGM1) in human cancer
- PMID: 38472489
- PMCID: PMC10933153
- DOI: 10.1007/s00432-024-05640-6
A pan-cancer analysis of the oncogenic and immunological roles of transglutaminase 1 (TGM1) in human cancer
Abstract
Background: There is currently a limited number of studies on transglutaminase type 1 (TGM1) in tumors. The objective of this study is to perform a comprehensive analysis across various types of cancer to determine the prognostic significance of TGM1 in tumors and investigate its role in the immune environment.
Method: Pan-cancer and mutational data were retrieved from the TCGA database and analyzed using R (version 3.6.4) and its associated software package. The expression difference and prognosis of TGM1 were examined, along with its correlation with tumor heterogeneity, stemness, mutation landscape, and RNA modification. Additionally, the relationship between TGM1 expression and tumor immunity was investigated using the TIMER method.
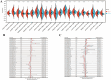
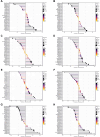
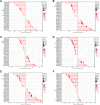
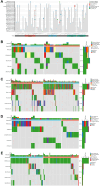
Results: TGM1 is expressed differently in various tumors and normal samples and is associated with the overall survival and progression-free time of KIRC, ACC, SKCM, LIHC, and STES. In LICH, we found a negative correlation between TGM1 expression and 6 indicators of tumor stemness. The mutation frequencies of BLCA, LIHC, and KIRC were 1.7%, 0.3%, and 0.3% respectively. In BLCA and BRCA, there was a significant correlation between TGM1 expression and the infiltration of CD4 + T cells, CD8 + T cells, neutrophils, and dendritic cells.
Conclusion: TGM1 has the potential to serve as both a prognostic marker and a drug target.
Keywords: Pan-cancer biomarker; Transglutaminase 1; Tumor-infiltrating cells.
© 2024. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
A pan-cancer analysis of the oncogenic role of zinc finger protein 419 in human cancer.Front Oncol. 2022 Dec 12;12:1042118. doi: 10.3389/fonc.2022.1042118. eCollection 2022. Front Oncol. 2022. PMID: 36578929 Free PMC article.
-
Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures.Front Immunol. 2021 Aug 31;12:721030. doi: 10.3389/fimmu.2021.721030. eCollection 2021. Front Immunol. 2021. PMID: 34531868 Free PMC article.
-
A pan-cancer analysis of the oncogenic and immunological roles of apolipoprotein F (APOF) in human cancer.Eur J Med Res. 2023 Jun 14;28(1):190. doi: 10.1186/s40001-023-01156-w. Eur J Med Res. 2023. PMID: 37312170 Free PMC article.
-
[Pan-cancer analysis of ubiquitin-specific protease 7 and its expression changes in the carcinogenesis of scar ulcer].Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2023 Jun 20;39(6):518-526. doi: 10.3760/cma.j.cn501225-20230421-00137. Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2023. PMID: 37805766 Free PMC article. Chinese.
-
Pan-cancer analysis of prognostic and immunological role of DTYMK in human tumors.Front Genet. 2022 Sep 8;13:989460. doi: 10.3389/fgene.2022.989460. eCollection 2022. Front Genet. 2022. PMID: 36159971 Free PMC article.
Cited by
-
Transglutaminase 1: Emerging Functions beyond Skin.Int J Mol Sci. 2024 Sep 25;25(19):10306. doi: 10.3390/ijms251910306. Int J Mol Sci. 2024. PMID: 39408635 Free PMC article. Review.
-
Exploring the role of ADAMTSL2 across multiple cancer types: A pan-cancer analysis and validated in colorectal cancer.Discov Oncol. 2024 Oct 9;15(1):538. doi: 10.1007/s12672-024-01401-6. Discov Oncol. 2024. PMID: 39384622 Free PMC article.
-
RAP1GAP is a prognostic biomarker and correlates with immune infiltrates in bladder cancer.Discov Oncol. 2025 May 22;16(1):863. doi: 10.1007/s12672-025-02634-9. Discov Oncol. 2025. PMID: 40405009 Free PMC article.
References
-
- Andersen PK, Gill RD (1982) Cox’s regression model for counting processes: a large sample study. Ann Stat. 10.1214/aos/1176345976
-
- Awadalla A et al (2022) Prognostic influence of microsatellite alterations of muscle-invasive bladder cancer treated with radical cystectomy. Urol Oncol 40(2):64.e9-64.e15 - PubMed
-
- Bianchi N, Beninati S, Bergamini CM (2018) Spotlight on the transglutaminase 2 gene: a focus on genomic and transcriptional aspects. Biochem J 475(9):1643–1667 - PubMed
-
- Blackhall F, Cappuzzo F, Crizotinib: from discovery to accelerated development to front-line treatment. Ann Oncol, 2016. 27 Suppl 3: p. iii35-iii41. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

