Investigating the Role of OrbF in Biofilm Biosynthesis and Regulation of Biofilm-Associated Genes in Bacillus cereus BC1
- PMID: 38472751
- PMCID: PMC10931033
- DOI: 10.3390/foods13050638
Investigating the Role of OrbF in Biofilm Biosynthesis and Regulation of Biofilm-Associated Genes in Bacillus cereus BC1
Abstract
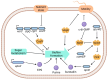
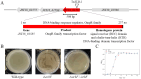
Bacillus cereus (B. cereus), a prevalent foodborne pathogen, constitutes a substantial risk to food safety due to its pronounced resilience under adverse environmental conditions such as elevated temperatures and ultraviolet radiation. This resilience can be attributed to its capacity for biofilm synthesis and sustained high viability. Our research aimed to elucidate the mechanisms governing biofilm biosynthesis in B. cereus. To this end, we constructed a 5088-mutant library of the B. cereus strain BC1 utilizing the transposon TnYLB-1. Systematic screening of this library yielded mutants exhibiting diminished biofilm formation capabilities. Twenty-four genes associated with the biofilm synthesis were identified by reverse PCR in these mutants, notably revealing a significant reduction in biofilm synthesis upon disruption of the orbF gene in B. cereus BC1. Comparative analysis between the wild type and orbF-deficient BC1 strains (BC1ΔorbF) indicated a marked downregulation (decreased by 11.7% to 96.7%) in the expression of genes implicated in biofilm formation, flagellar assembly, and bacterial chemotaxis in the BC1ΔorbF. Electrophoretic mobility shift assay (EMSA) further corroborated the role of OrbF, demonstrating its binding to the promoter region of the biofilm gene cluster, subsequently leading to the suppression of transcriptional activity of biofilm-associated genes in B. cereus BC1. Our findings underscore the pivotal role of orbF in biofilm biosynthesis in B. cereus, highlighting its potential as a target for strategies aimed at mitigating biofilm formation in this pathogen.
Keywords: Bacillus cereus; biofilm formation; regulator OrbF; regulatory mechanism; transposon TnYLB-1.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
SpoVG is an important regulator of sporulation and affects biofilm formation by regulating Spo0A transcription in Bacillus cereus 0-9.BMC Microbiol. 2021 Jun 8;21(1):172. doi: 10.1186/s12866-021-02239-6. BMC Microbiol. 2021. PMID: 34102998 Free PMC article.
-
A transposon mutant library of Bacillus cereus ATCC 10987 reveals novel genes required for biofilm formation and implicates motility as an important factor for pellicle-biofilm formation.Microbiologyopen. 2018 Apr;7(2):e00552. doi: 10.1002/mbo3.552. Epub 2017 Nov 22. Microbiologyopen. 2018. PMID: 29164822 Free PMC article.
-
A Flagella Hook Coding Gene flgE Positively Affects Biofilm Formation and Cereulide Production in Emetic Bacillus cereus.Front Microbiol. 2022 Jun 10;13:897836. doi: 10.3389/fmicb.2022.897836. eCollection 2022. Front Microbiol. 2022. PMID: 35756067 Free PMC article.
-
Genome-Wide Investigation of Biofilm Formation in Bacillus cereus.Appl Environ Microbiol. 2017 Jun 16;83(13):e00561-17. doi: 10.1128/AEM.00561-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28432092 Free PMC article.
-
Bacillus cereus spores and toxins - The potential role of biofilms.Food Microbiol. 2020 Sep;90:103493. doi: 10.1016/j.fm.2020.103493. Epub 2020 Mar 26. Food Microbiol. 2020. PMID: 32336372 Review.
Cited by
-
Essential oils and Lactobacillus metabolites as alternative antibiofilm agents against foodborne bacteria and molecular analysis of biofilm regulatory genes.Sci Rep. 2025 Mar 4;15(1):7576. doi: 10.1038/s41598-025-89998-8. Sci Rep. 2025. PMID: 40038354 Free PMC article.
References
-
- Gao T., Ding Y., Wu Q., Wang J., Zhang J., Yu S., Yu P., Liu C., Kong L., Feng Z., et al. Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus cereus Isolated from Pasteurized Milk in China. Front. Microbiol. 2018;9:533. doi: 10.3389/fmicb.2018.00533. - DOI - PMC - PubMed
Grants and funding
- 2023YFD2100500/National Key Research and Development Program of China
- U2106228/National Natural Science Foundation of China
- 32300059/National Natural Science Foundation of China
- XTC2205/the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture
- BK20230314/Natural Science Foundation of Jiangsu Province
LinkOut - more resources
Full Text Sources

