Plant and Arthropod IgE-Binding Papain-like Cysteine Proteases: Multiple Contributions to Allergenicity
- PMID: 38472904
- PMCID: PMC10931198
- DOI: 10.3390/foods13050790
Plant and Arthropod IgE-Binding Papain-like Cysteine Proteases: Multiple Contributions to Allergenicity
Abstract
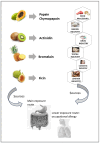
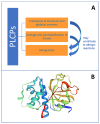
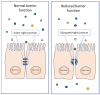
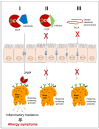
Papain-like cysteine proteases are widespread and can be detected in all domains of life. They share structural and enzymatic properties with the group's namesake member, papain. They show a broad range of protein substrates and are involved in several biological processes. These proteases are widely exploited for food, pharmaceutical, chemical and cosmetic biotechnological applications. However, some of them are known to cause allergic reactions. In this context, the objective of this review is to report an overview of some general properties of papain-like cysteine proteases and to highlight their contributions to allergy reactions observed in humans. For instance, the literature shows that their proteolytic activity can cause an increase in tissue permeability, which favours the crossing of allergens through the skin, intestinal and respiratory barriers. The observation that allergy to PLCPs is mostly detected for inhaled proteins is in line with the reports describing mite homologs, such as Der p 1 and Der f 1, as major allergens showing a frequent correlation between sensitisation and clinical allergic reactions. In contrast, the plant food homologs are often digested in the gastrointestinal tract. Therefore, they only rarely can cause allergic reactions in humans. Accordingly, they are reported mainly as a cause of occupational diseases.
Keywords: allergen homologs; gastrointestinal digestion; inhaled allergens; mite proteases; occupational allergy; plant food; proteolytic activity; sensitisation; tight junction; tissue permeabilisation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

