Preclinical Characterization of the Anti-Leukemia Activity of the CD33/CD16a/NKG2D Immune-Modulating TriNKET® CC-96191
- PMID: 38473239
- PMCID: PMC10931532
- DOI: 10.3390/cancers16050877
Preclinical Characterization of the Anti-Leukemia Activity of the CD33/CD16a/NKG2D Immune-Modulating TriNKET® CC-96191
Abstract
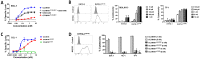
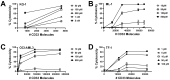
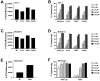
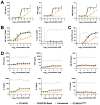
Increasing efforts are focusing on natural killer (NK) cell immunotherapies for AML. Here, we characterized CC-96191, a novel CD33/CD16a/NKG2D immune-modulating TriNKET®. CC-96191 simultaneously binds CD33, NKG2D, and CD16a, with NKG2D and CD16a co-engagement increasing the avidity for, and activation of, NK cells. CC-96191 was broadly active against human leukemia cells in a strictly CD33-dependent manner, with maximal efficacy requiring the co-engagement of CD16a and NKG2D. A frequent CD33 single nucleotide polymorphism, R69G, reduced CC-96191 potency but not maximal activity, likely because of reduced CD33 binding. Similarly, the potency, but not the maximal activity, of CC-96191 was reduced by high concentrations of soluble CD33; in contrast, the soluble form of the NKG2D ligand MICA did not impact activity. In the presence of CD33+ AML cells, CC-96191 activated NK cells but not T cells; while maximum anti-AML efficacy was similar, soluble cytokine levels were 10- to >100-fold lower than with a CD33/CD3 bispecific antibody. While CC-96191-mediated cytolysis was not affected by ABC transporter proteins, it was reduced by anti-apoptotic BCL-2 family proteins. Finally, in patient marrow specimens, CC-96191 eliminated AML cells but not normal monocytes, suggesting selectivity of TriNKET-induced cytotoxicity toward neoplastic cells. Together, these findings support the clinical exploration of CC-96191 as in NCT04789655.
Keywords: CD33; acute myeloid leukemia (AML); immunotherapy; natural killer (NK) cell; trispecific antibody.
Conflict of interest statement
M.T.O., H.C., H.J.T., H.K.J, M.A.J., and J.L. are employees of Bristol Myers Squibb. A.F.C, G.P.C, T.D., D.F., and A.G. are employees of Dragonfly Therapeutics. E.R.A. received honoraria for educational activities and/or conferences and/or travel grants from AbbVie, Astellas, Gilead, and Jazz. R.B.W. received laboratory research grants and/or clinical trial support from Aptevo, Celgene/Bristol Myers Squibb, ImmunoGen, Janssen, Jazz, Kite, Kura, Pfizer, and VOR; has ownership interests in Amphivena; and is (or has been) a consultant to Abbvie, Adicet, Amphivena, BerGenBio, Celgene/Bristol Myers Squibb, GlaxoSmithKline, ImmunoGen, Kura, Orum, and Wugen. The other authors declare no competing financial interests.
Figures


References
-
- Döhner H., Wei A.H., Appelbaum F.R., Craddock C., DiNardo C.D., Dombret H., Ebert B.L., Fenaux P., Godley L.A., Hasserjian R.P., et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–1377. doi: 10.1182/blood.2022016867. - DOI - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

