Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers
- PMID: 38473352
- PMCID: PMC10931536
- DOI: 10.3390/cancers16050991
Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers
Abstract
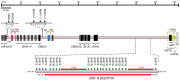
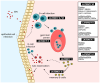
The Epstein-Barr Virus (EBV) is a double-stranded DNA-based human tumor virus that was first isolated in 1964 from lymphoma biopsies. Since its initial discovery, EBV has been identified as a major contributor to numerous cancers and chronic autoimmune disorders. The virus is particularly efficient at infecting B-cells but can also infect epithelial cells, utilizing an array of epigenetic strategies to establish long-term latent infection. The association with histone modifications, alteration of DNA methylation patterns in host and viral genomes, and microRNA targeting of host cell factors are core epigenetic strategies that drive interactions between host and virus, which are necessary for viral persistence and progression of EBV-associated diseases. Therefore, understanding epigenetic regulation and its role in post-entry viral dynamics is an elusive area of EBV research. Here, we present current outlooks of EBV epigenetic regulation as it pertains to viral interactions with its host during latent infection and its propensity to induce tumorigenesis. We review the important epigenetic regulators of EBV latency and explore how the strategies involved during latent infection drive differential epigenetic profiles and host-virus interactions in EBV-associated cancers.
Keywords: EBV-associated cancers; Epstein–Barr virus; host virus interactions; microRNAs; tumor viruses; viral epigenetics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Rous P., Murphy J.B., Tytler W.H. A filterable agent the cause of a second chicken-tumor, an osteochondrosarcoma. J. Am. Med. Assoc. 1912;59:1793–1794. doi: 10.1001/jama.1912.04270110207011. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

