Transcriptomic, Proteomic, and Genomic Mutational Fraction Differences Based on HPV Status Observed in Patient-Derived Xenograft Models of Penile Squamous Cell Carcinoma
- PMID: 38473423
- PMCID: PMC10930474
- DOI: 10.3390/cancers16051066
Transcriptomic, Proteomic, and Genomic Mutational Fraction Differences Based on HPV Status Observed in Patient-Derived Xenograft Models of Penile Squamous Cell Carcinoma
Abstract
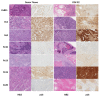
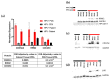
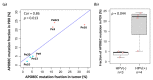
Metastatic penile squamous cell carcinoma (PSCC) has only a 50% response rate to first-line combination chemotherapies and there are currently no targeted-therapy approaches. Therefore, we have an urgent need in advanced-PSCC treatment to find novel therapies. Approximately half of all PSCC cases are positive for high-risk human papillomavirus (HR-HPV). Our objective was to generate HPV-positive (HPV+) and HPV-negative (HPV-) patient-derived xenograft (PDX) models and to determine the biological differences between HPV+ and HPV- disease. We generated four HPV+ and three HPV- PSCC PDX animal models by directly implanting resected patient tumor tissue into immunocompromised mice. PDX tumor tissue was found to be similar to patient tumor tissue (donor tissue) by histology and short tandem repeat fingerprinting. DNA mutations were mostly preserved in PDX tissues and similar APOBEC (apolipoprotein B mRNA editing catalytic polypeptide) mutational fractions in donor tissue and PDX tissues were noted. A higher APOBEC mutational fraction was found in HPV+ versus HPV- PDX tissues (p = 0.044), and significant transcriptomic and proteomic expression differences based on HPV status included p16 (CDKN2A), RRM2, and CDC25C. These models will allow for the direct testing of targeted therapies in PSCC and determine their response in correlation to HPV status.
Keywords: APOBEC mutations; human papillomavirus-positive penile squamous cell carcinoma; patient-derived xenograft; penile squamous cell carcinoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Establishment and Characterization of Advanced Penile Cancer Patient-derived Tumor Xenografts: Paving the Way for Personalized Treatments.Eur Urol Focus. 2022 Nov;8(6):1787-1794. doi: 10.1016/j.euf.2022.04.012. Epub 2022 May 7. Eur Urol Focus. 2022. PMID: 35537937
-
Comprehensive genomic profiling of penile squamous cell carcinoma and the impact of human papillomavirus status on immune-checkpoint inhibitor-related biomarkers.Cancer. 2023 Dec 15;129(24):3884-3893. doi: 10.1002/cncr.34982. Epub 2023 Aug 11. Cancer. 2023. PMID: 37565840
-
Single-cell Atlas of Penile Cancer Reveals TP53 Mutations as a Driver of an Aggressive Phenotype, Irrespective of Human Papillomavirus Status, and Provides Clues for Treatment Personalization.Eur Urol. 2024 Aug;86(2):114-127. doi: 10.1016/j.eururo.2024.03.038. Epub 2024 Apr 26. Eur Urol. 2024. PMID: 38670879
-
The Prognostic Role of Human Papillomavirus and p16 Status in Penile Squamous Cell Carcinoma-A Systematic Review.Cancers (Basel). 2023 Jul 21;15(14):3713. doi: 10.3390/cancers15143713. Cancers (Basel). 2023. PMID: 37509374 Free PMC article. Review.
-
Pathogenesis of Penile Squamous Cell Carcinoma: Molecular Update and Systematic Review.Int J Mol Sci. 2021 Dec 27;23(1):251. doi: 10.3390/ijms23010251. Int J Mol Sci. 2021. PMID: 35008677 Free PMC article.
Cited by
-
Differential Efficacy of Bevacizumab and Erlotinib in Preclinical Models of Renal Medullary Carcinoma and Fumarate Hydratase-Deficient Renal Cell Carcinoma.Mol Cancer Ther. 2025 Jul 2:10.1158/1535-7163.MCT-24-0703. doi: 10.1158/1535-7163.MCT-24-0703. Online ahead of print. Mol Cancer Ther. 2025. PMID: 40601845 Free PMC article.
References
-
- Steinestel J., Al Ghazal A., Arndt A., Schnoeller T.J., Schrader A.J., Moeller P., Steinestel K. The role of histologic subtype, p16(INK4a) expression, and presence of human papillomavirus DNA in penile squamous cell carcinoma. BMC Cancer. 2015;15:220. doi: 10.1186/s12885-015-1268-z. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

