Current Strategies for Increasing Knock-In Efficiency in CRISPR/Cas9-Based Approaches
- PMID: 38473704
- PMCID: PMC10931195
- DOI: 10.3390/ijms25052456
Current Strategies for Increasing Knock-In Efficiency in CRISPR/Cas9-Based Approaches
Abstract
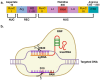
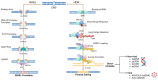
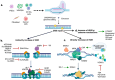
Since its discovery in 2012, the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system has supposed a promising panorama for developing novel and highly precise genome editing-based gene therapy (GT) alternatives, leading to overcoming the challenges associated with classical GT. Classical GT aims to deliver transgenes to the cells via their random integration in the genome or episomal persistence into the nucleus through lentivirus (LV) or adeno-associated virus (AAV), respectively. Although high transgene expression efficiency is achieved by using either LV or AAV, their nature can result in severe side effects in humans. For instance, an LV (NCT03852498)- and AAV9 (NCT05514249)-based GT clinical trials for treating X-linked adrenoleukodystrophy and Duchenne Muscular Dystrophy showed the development of myelodysplastic syndrome and patient's death, respectively. In contrast with classical GT, the CRISPR/Cas9-based genome editing requires the homologous direct repair (HDR) machinery of the cells for inserting the transgene in specific regions of the genome. This sophisticated and well-regulated process is limited in the cell cycle of mammalian cells, and in turn, the nonhomologous end-joining (NHEJ) predominates. Consequently, seeking approaches to increase HDR efficiency over NHEJ is crucial. This manuscript comprehensively reviews the current alternatives for improving the HDR for CRISPR/Cas9-based GTs.
Keywords: CRISPR/Cas9; HDR; NHEJ; genome editing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks.Int J Mol Sci. 2020 Sep 4;21(18):6461. doi: 10.3390/ijms21186461. Int J Mol Sci. 2020. PMID: 32899704 Free PMC article. Review.
-
Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish.Methods. 2017 May 15;121-122:77-85. doi: 10.1016/j.ymeth.2017.03.005. Epub 2017 Mar 12. Methods. 2017. PMID: 28300641 Review.
-
Precision genome editing in the CRISPR era.Biochem Cell Biol. 2017 Apr;95(2):187-201. doi: 10.1139/bcb-2016-0137. Epub 2016 Sep 29. Biochem Cell Biol. 2017. PMID: 28177771 Review.
-
CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites.Nat Commun. 2019 Jun 28;10(1):2866. doi: 10.1038/s41467-019-10735-7. Nat Commun. 2019. PMID: 31253785 Free PMC article.
-
A high-efficiency and versatile CRISPR/Cas9-mediated HDR-based biallelic editing system.J Zhejiang Univ Sci B. 2022 Feb 15;23(2):141-152. doi: 10.1631/jzus.B2100196. J Zhejiang Univ Sci B. 2022. PMID: 35187887 Free PMC article. English.
Cited by
-
The Role of Somatic Cell Synchronization in Nuclear Transfer and Induced Pluripotent Stem Cells for Wild Felids.Zoo Biol. 2025 May-Jun;44(3):221-229. doi: 10.1002/zoo.21896. Epub 2025 Mar 7. Zoo Biol. 2025. PMID: 40051311 Free PMC article. Review.
-
Gene therapy for genetic diseases: challenges and future directions.MedComm (2020). 2025 Feb 13;6(2):e70091. doi: 10.1002/mco2.70091. eCollection 2025 Feb. MedComm (2020). 2025. PMID: 39949979 Free PMC article. Review.
-
CRISPR-Cas Systems and Genome Editing: Beginning the Era of CRISPR/Cas Therapies for Humans.Int J Mol Sci. 2024 May 13;25(10):5292. doi: 10.3390/ijms25105292. Int J Mol Sci. 2024. PMID: 38791336 Free PMC article.
-
Strategies for CRISPR-based knock-ins in primary human B cells and lymphoma cell lines.Front Immunol. 2025 May 16;16:1589729. doi: 10.3389/fimmu.2025.1589729. eCollection 2025. Front Immunol. 2025. PMID: 40453079 Free PMC article. Review.
-
Assessing Microprocessor complex mutations with a Microsensor system.RNA. 2025 Jun 16;31(7):896-915. doi: 10.1261/rna.080338.124. RNA. 2025. PMID: 40328469
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

