Plasma Lipidomic Profiling Using Mass Spectrometry for Multiple Sclerosis Diagnosis and Disease Activity Stratification (LipidMS)
- PMID: 38473733
- PMCID: PMC10932002
- DOI: 10.3390/ijms25052483
Plasma Lipidomic Profiling Using Mass Spectrometry for Multiple Sclerosis Diagnosis and Disease Activity Stratification (LipidMS)
Abstract
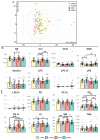
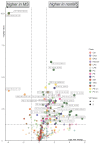
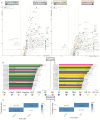
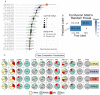
This investigation explores the potential of plasma lipidomic signatures for aiding in the diagnosis of Multiple Sclerosis (MS) and evaluating the clinical course and disease activity of diseased patients. Plasma samples from 60 patients with MS (PwMS) were clinically stratified to either a relapsing-remitting (RRMS) or a chronic progressive MS course and 60 age-matched controls were analyzed using state-of-the-art direct infusion quantitative shotgun lipidomics. To account for potential confounders, data were filtered for age and BMI correlations. The statistical analysis employed supervised and unsupervised multivariate data analysis techniques, including a principal component analysis (PCA), a partial least squares discriminant analysis (oPLS-DA) and a random forest (RF). To determine whether the significant absolute differences in the lipid subspecies have a relevant effect on the overall composition of the respective lipid classes, we introduce a class composition visualization (CCV). We identified 670 lipids across 16 classes. PwMS showed a significant increase in diacylglycerols (DAG), with DAG 16:0;0_18:1;0 being proven to be the lipid with the highest predictive ability for MS as determined by RF. The alterations in the phosphatidylethanolamines (PE) were mainly linked to RRMS while the alterations in the ether-bound PEs (PE O-) were found in chronic progressive MS. The amount of CE species was reduced in the CPMS cohort whereas TAG species were reduced in the RRMS patients, both lipid classes being relevant in lipid storage. Combining the above mentioned data analyses, distinct lipidomic signatures were isolated and shown to be correlated with clinical phenotypes. Our study suggests that specific plasma lipid profiles are not merely associated with the diagnosis of MS but instead point toward distinct clinical features in the individual patient paving the way for personalized therapy and an enhanced understanding of MS pathology.
Keywords: biomarker; lipidmetabolism; lipidomics; multiple sclerosis.
Conflict of interest statement
S.S.J.L., L.-M.B., K.a.d.B., L.V., M.N. and D.F. declare no conflicts of interest. C.K. is an employee and shareholder of Lipotype GmbH, Dresden.
Figures

References
-
- Wallin M.T., Culpepper W.J., Nichols E., Bhutta Z.A., Gebrehiwot T.T., Hay S.I., Khalil I.A., Krohn K.J., Liang X., Naghavi M., et al. Global, Regional, and National Burden of Multiple Sclerosis 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:269–285. doi: 10.1016/S1474-4422(18)30443-5. - DOI - PMC - PubMed
-
- Villoslada P., Alonso C., Agirrezabal I., Kotelnikova E., Zubizarreta I., Pulido-Valdeolivas I., Saiz A., Comabella M., Montalban X., Villar L., et al. Metabolomic Signatures Associated with Disease Severity in Multiple Sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2017;4:e321. doi: 10.1212/NXI.0000000000000321. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

