Unlocking the Transcriptional Control of NCAPG in Bovine Myoblasts: CREB1 and MYOD1 as Key Players
- PMID: 38473754
- PMCID: PMC10931257
- DOI: 10.3390/ijms25052506
Unlocking the Transcriptional Control of NCAPG in Bovine Myoblasts: CREB1 and MYOD1 as Key Players
Abstract
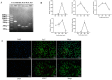
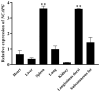
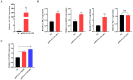
Muscle formation directly determines meat production and quality. The non-SMC condensin I complex subunit G (NCAPG) is strongly linked to the growth features of domestic animals because it is essential in controlling muscle growth and development. This study aims to elucidate the tissue expression level of the bovine NCAPG gene, and determine the key transcription factors for regulating the bovine NCAPG gene. In this study, we observed that the bovine NCAPG gene exhibited high expression levels in longissimus dorsi and spleen tissues. Subsequently, we cloned and characterized the promoter region of the bovine NCAPG gene, consisting of a 2039 bp sequence, through constructing the deletion fragment double-luciferase reporter vector and site-directed mutation-identifying core promoter region with its key transcription factor binding site. In addition, the key transcription factors of the core promoter sequence of the bovine NCAPG gene were analyzed and predicted using online software. Furthermore, by integrating overexpression experiments and the electrophoretic mobility shift assay (EMSA), we have shown that cAMP response element binding protein 1 (CREB1) and myogenic differentiation 1 (MYOD1) bind to the core promoter region (-598/+87), activating transcription activity in the bovine NCAPG gene. In conclusion, these findings shed important light on the regulatory network mechanism that underlies the expression of the NCAPG gene throughout the development of the muscles in beef cattle.
Keywords: NCAPG; bovine; myoblast; promoter; transcriptional regulation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
- GAU-QDFC-2022-07/Science and Technology Innovation Funds of Gansu Agricultural University-Supporting funds for youth mentor
- KJJC-LX-2023-3-01/Integration and demonstration of key technologies for healthy and efficient breeding of beef cows in Linxia
- GAU-XKTD-2022-22/Discipline Team Project of Gansu Agricultural University
- 32360821/the National Natural Science Foundation
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

