Multi-Omics Analysis Reveals That Anthocyanin Degradation and Phytohormone Changes Regulate Red Color Fading in Rapeseed (Brassica napus L.) Petals
- PMID: 38473825
- PMCID: PMC10931936
- DOI: 10.3390/ijms25052577
Multi-Omics Analysis Reveals That Anthocyanin Degradation and Phytohormone Changes Regulate Red Color Fading in Rapeseed (Brassica napus L.) Petals
Abstract
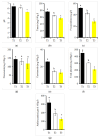
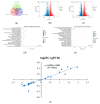
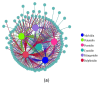
Flower color is an important trait for the ornamental value of colored rapeseed (Brassica napus L.), as the plant is becoming more popular. However, the color fading of red petals of rapeseed is a problem for its utilization. Unfortunately, the mechanism for the process of color fading in rapeseed is unknown. In the current study, a red flower line, Zhehuhong, was used as plant material to analyze the alterations in its morphological and physiological characteristics, including pigment and phytohormone content, 2 d before flowering (T1), at flowering (T2), and 2 d after flowering (T3). Further, metabolomics and transcriptomics analyses were also performed to reveal the molecular regulation of petal fading. The results show that epidermal cells changed from spherical and tightly arranged to totally collapsed from T1 to T3, according to both paraffin section and scanning electron microscope observation. The pH value and all pigment content except flavonoids decreased significantly during petal fading. The anthocyanin content was reduced by 60.3% at T3 compared to T1. The content of three phytohormones, 1-aminocyclopropanecarboxylic acid, melatonin, and salicylic acid, increased significantly by 2.2, 1.1, and 30.3 times, respectively, from T1 to T3. However, auxin, abscisic acid, and jasmonic acid content decreased from T1 to T3. The result of metabolomics analysis shows that the content of six detected anthocyanin components (cyanidin, peonidin, pelargonidin, delphinidin, petunidin, and malvidin) and their derivatives mainly exhibited a decreasing trend, which was in accordance with the trend of decreasing anthocyanin. Transcriptomics analysis showed downregulation of genes involved in flavonol, flavonoid, and anthocyanin biosynthesis. Furthermore, genes regulating anthocyanin biosynthesis were preferentially expressed at early stages, indicating that the degradation of anthocyanin is the main issue during color fading. The corresponding gene-encoding phytohormone biosynthesis and signaling, JASMONATE-ZIM-DOMAIN PROTEIN, was deactivated to repress anthocyanin biosynthesis, resulting in fading petal color. The results clearly suggest that anthocyanin degradation and phytohormone regulation play essential roles in petal color fading in rapeseed, which is a useful insight for the breeding of colored rapeseed.
Keywords: Brassica napus L.; anthocyanin; fading; metabolomics; petal; phytohormone; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- Fu D.H., Jiang L.Y., Mason A.S., Xiao M.L., Zhu L.R., Li L.Z., Zhou Q.H., Shen C.J., Huang C.H. Research progress and strategies for multifunctional rapeseed: A case study of China. J. Integr. Agric. 2016;15:1673–1684. doi: 10.1016/S2095-3119(16)61384-9. - DOI
-
- Yin N.W., Wang S.X., Jia L.D., Zhu M.C., Yang J., Zhou B.J., Yin J.M., Lu K., Wang R., Li J.N., et al. Identification and Characterization of Major Constituents in Different-Colored Rapeseed Petals by UPLC-HESI-MS/MS. Agric. Food Chem. 2019;67:11053–11065. doi: 10.1021/acs.jafc.9b05046. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

