The Microtubule End Binding Protein Mal3 Is Essential for the Dynamic Assembly of Microtubules during Magnaporthe oryzae Growth and Pathogenesis
- PMID: 38473921
- PMCID: PMC10932144
- DOI: 10.3390/ijms25052672
The Microtubule End Binding Protein Mal3 Is Essential for the Dynamic Assembly of Microtubules during Magnaporthe oryzae Growth and Pathogenesis
Abstract
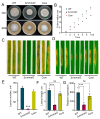
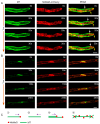
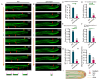
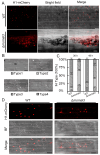
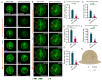
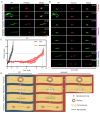
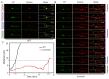
Cytoskeletal microtubules (MTs) play crucial roles in many aspects of life processes in eukaryotic organisms. They dynamically assemble physiologically important MT arrays under different cell conditions. Currently, aspects of MT assembly underlying the development and pathogenesis of the model plant pathogenic fungus Magnaporthe oryzae (M. oryzae) are unclear. In this study, we characterized the MT plus end binding protein MoMal3 in M. oryzae. We found that knockout of MoMal3 results in defects in hyphal polar growth, appressorium-mediated host penetration and nucleus division. Using high-resolution live-cell imaging, we further found that the MoMal3 mutant assembled a rigid MT in parallel with the MT during hyphal polar growth, the cage-like network in the appressorium and the stick-like spindle in nuclear division. These aberrant MT organization patterns in the MoMal3 mutant impaired actin-based cell growth and host infection. Taken together, these findings showed that M. oryzae relies on MoMal3 to assemble elaborate MT arrays for growth and infection. The results also revealed the assembly mode of MTs in M. oryzae, indicating that MTs are pivotal for M. oryzae growth and host infection and may be new targets for devastating fungus control.
Keywords: Magnaporthe oryzae; dynamic assembly; infection mechanism; microtubule cytoskeleton; microtubule plus end; nucleus division; rice blast.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Bacillus subtilis KLBMPGC81 suppresses appressorium-mediated plant infection by altering the cell wall integrity signaling pathway and multiple cell biological processes in Magnaporthe oryzae.Front Cell Infect Microbiol. 2022 Sep 9;12:983757. doi: 10.3389/fcimb.2022.983757. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36159636 Free PMC article.
-
Nucleosome Assembly Protein 1, Nap1, Is Required for the Growth, Development, and Pathogenicity of Magnaporthe oryzae.Int J Mol Sci. 2022 Jul 11;23(14):7662. doi: 10.3390/ijms23147662. Int J Mol Sci. 2022. PMID: 35887015 Free PMC article.
-
The MoLfa1 Protein Regulates Fungal Development and Septin Ring Formation in Magnaporthe oryzae.Int J Mol Sci. 2024 Mar 19;25(6):3434. doi: 10.3390/ijms25063434. Int J Mol Sci. 2024. PMID: 38542408 Free PMC article.
-
Investigating the cell and developmental biology of plant infection by the rice blast fungus Magnaporthe oryzae.Fungal Genet Biol. 2021 Sep;154:103562. doi: 10.1016/j.fgb.2021.103562. Epub 2021 Apr 18. Fungal Genet Biol. 2021. PMID: 33882359 Review.
-
Effector-triggered susceptibility by the rice blast fungus Magnaporthe oryzae.New Phytol. 2024 Feb;241(3):1007-1020. doi: 10.1111/nph.19446. Epub 2023 Dec 10. New Phytol. 2024. PMID: 38073141 Review.
References
-
- Eseola A.B., Ryder L.S., Oses-Ruiz M., Findlay K., Yan X., Cruz-Mireles N., Molinari C., Garduno-Rosales M., Talbot N.J. Investigating the cell and developmental biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2021;154:103562. doi: 10.1016/j.fgb.2021.103562. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

