Similarities in Structure and Function of UDP-Glycosyltransferase Homologs from Human and Plants
- PMID: 38474028
- PMCID: PMC10932239
- DOI: 10.3390/ijms25052782
Similarities in Structure and Function of UDP-Glycosyltransferase Homologs from Human and Plants
Abstract
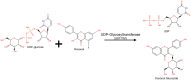
The uridine diphosphate glycosyltransferase (UGT) superfamily plays a key role in the metabolism of xenobiotics and metabolic wastes, which is essential for detoxifying those species. Over the last several decades, a huge effort has been put into studying human and mammalian UGT homologs, but family members in other organisms have been explored much less. Potentially, other UGT homologs can have desirable substrate specificity and biological activities that can be harnessed for detoxification in various medical settings. In this review article, we take a plant UGT homology, UGT71G1, and compare its structural and biochemical properties with the human homologs. These comparisons suggest that even though mammalian and plant UGTs are functional in different environments, they may support similar biochemical activities based on their protein structure and function. The known biological functions of these homologs are discussed so as to provide insights into the use of UGT homologs from other organisms for addressing human diseases related to UGTs.
Keywords: UGT-related diseases; glycosylation; substrate specificity; uridine diphosphate glycosyltransferases.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

