Circ MTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3
- PMID: 38474064
- PMCID: PMC10932366
- DOI: 10.3390/ijms25052817
Circ MTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3
Abstract
Our previous study has reported that metastasis-associated protein 2 (MTA2) plays essential roles in tumorigenesis and aggressiveness of gastric cancer (GC). However, the underlying molecular mechanisms of MTA2-mediated GC and its upstream regulation mechanism remain elusive. In this study, we identified a novel circular RNA (circRNA) generated from the MTA2 gene (circMTA2) as a crucial regulator in GC progression. CircMTA2 was highly expressed in GC tissues and cell lines, and circMTA2 promoted the proliferation, invasion, and metastasis of GC cells both in vitro and in vivo. Mechanistically, circMTA2 interacted with ubiquitin carboxyl-terminal hydrolase L3 (UCHL3) to restrain MTA2 ubiquitination and stabilize MTA2 protein expression, thereby facilitating tumor progression. Moreover, circMTA2 was mainly encapsulated and transported by exosomes to promote GC cell progression. Taken together, these findings uncover that circMTA2 suppresses MTA2 degradation by interacting with UCHL3, thereby promoting GC progression. In conclusion, we identified a cancer-promoting axis (circMTA2/UCHL3/MTA2) in GC progression, which paves the way for us to design and synthesize targeted inhibitors as well as combination therapies.
Keywords: CircRNAs; MTA2; UCHL3; gastric cancer; ubiquitination.
Conflict of interest statement
The authors have no conflict of interest.
Figures

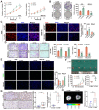

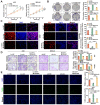
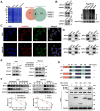

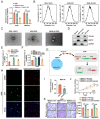
References
-
- Kakeji Y., Ishikawa T., Suzuki S., Akazawa K., Irino T., Miyashiro I., Ono H., Suzuki H., Tanabe S., Kadowaki S., et al. A retrospective 5-year survival analysis of surgically resected gastric cancer cases from the Japanese Gastric Cancer Association nationwide registry (2001–2013) Gastric Cancer. 2022;25:1082–1093. doi: 10.1007/s10120-022-01317-6. - DOI - PubMed
-
- Chung H., Ko Y., Lee I.S., Hur H., Huh J., Han S.U., Kim K.W., Lee J. Prognostic artificial intelligence model to predict 5 year survival at 1 year after gastric cancer surgery based on nutrition and body morphometry. J. Cachexia Sarcopenia Muscle. 2023;14:847–859. doi: 10.1002/jcsm.13176. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

