Development of Fluorescence-Based Assays for Key Viral Proteins in the SARS-CoV-2 Infection Process and Lifecycle
- PMID: 38474097
- PMCID: PMC10932265
- DOI: 10.3390/ijms25052850
Development of Fluorescence-Based Assays for Key Viral Proteins in the SARS-CoV-2 Infection Process and Lifecycle
Abstract
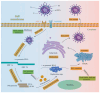
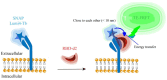
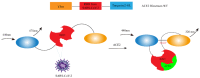
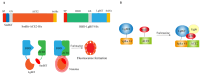
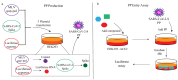
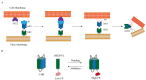
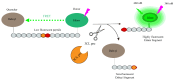
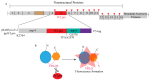
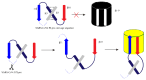
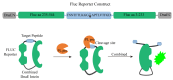
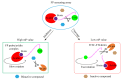
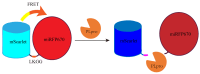
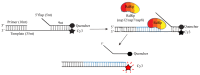
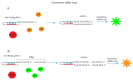
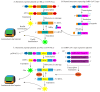
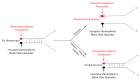
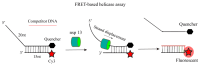
Since the appearance of SARS-CoV-2 in 2019, the ensuing COVID-19 (Corona Virus Disease 2019) pandemic has posed a significant threat to the global public health system, human health, life, and economic well-being. Researchers worldwide have devoted considerable efforts to curb its spread and development. The latest studies have identified five viral proteins, spike protein (Spike), viral main protease (3CLpro), papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp), and viral helicase (Helicase), which play crucial roles in the invasion of SARS-CoV-2 into the human body and its lifecycle. The development of novel anti-SARS-CoV-2 drugs targeting these five viral proteins holds immense promise. Therefore, the development of efficient, high-throughput screening methodologies specifically designed for these viral proteins is of utmost importance. Currently, a plethora of screening techniques exists, with fluorescence-based assays emerging as predominant contenders. In this review, we elucidate the foundational principles and methodologies underpinning fluorescence-based screening approaches directed at these pivotal viral targets, hoping to guide researchers in the judicious selection and refinement of screening strategies, thereby facilitating the discovery and development of lead compounds for anti-SARS-CoV-2 pharmaceuticals.
Keywords: COVID-19; SARS-CoV-2; antiviral agents; assay development; infection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


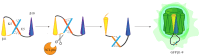


References
-
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

