Lack of the Histone Deacetylase SIRT1 Leads to Protection against Endoplasmic Reticulum Stress through the Upregulation of Heat Shock Proteins
- PMID: 38474102
- PMCID: PMC10932345
- DOI: 10.3390/ijms25052856
Lack of the Histone Deacetylase SIRT1 Leads to Protection against Endoplasmic Reticulum Stress through the Upregulation of Heat Shock Proteins
Abstract
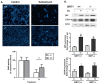
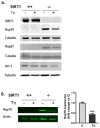
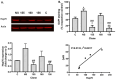
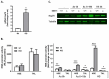
Histone deacetylase SIRT1 represses gene expression through the deacetylation of histones and transcription factors and is involved in the protective cell response to stress and aging. However, upon endoplasmic reticulum (ER) stress, SIRT1 impairs the IRE1α branch of the unfolded protein response (UPR) through the inhibition of the transcriptional activity of XBP-1 and SIRT1 deficiency is beneficial under these conditions. We hypothesized that SIRT1 deficiency may unlock the blockade of transcription factors unrelated to the UPR promoting the synthesis of chaperones and improving the stability of immature proteins or triggering the clearance of unfolded proteins. SIRT1+/+ and SIRT1-/- fibroblasts were exposed to the ER stress inducer tunicamycin and cell survival and expression of heat shock proteins were analyzed 24 h after the treatment. We observed that SIRT1 loss significantly reduced cell sensitivity to ER stress and showed that SIRT1-/- but not SIRT1+/+ cells constitutively expressed high levels of phospho-STAT3 and heat shock proteins. Hsp70 silencing in SIRT1-/- cells abolished the resistance to ER stress. Furthermore, accumulation of ubiquitinated proteins was lower in SIRT1-/- than in SIRT1+/+ cells. Our data showed that SIRT1 deficiency enabled chaperones upregulation and boosted the proteasome activity, two processes that are beneficial for coping with ER stress.
Keywords: ER stress; Hsp70; STAT3; sirtuin.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures
Similar articles
-
SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation.Cell Death Differ. 2017 Feb;24(2):343-356. doi: 10.1038/cdd.2016.138. Epub 2016 Dec 2. Cell Death Differ. 2017. PMID: 27911441 Free PMC article.
-
Effects of muscular dystrophy, exercise and blocking activin receptor IIB ligands on the unfolded protein response and oxidative stress.Free Radic Biol Med. 2016 Oct;99:308-322. doi: 10.1016/j.freeradbiomed.2016.08.017. Epub 2016 Aug 20. Free Radic Biol Med. 2016. PMID: 27554968
-
Quercetin affects Hsp70/IRE1α mediated protection from death induced by endoplasmic reticulum stress.Oxid Med Cell Longev. 2015;2015:645157. doi: 10.1155/2015/645157. Epub 2015 Apr 2. Oxid Med Cell Longev. 2015. PMID: 25922642 Free PMC article.
-
Molecular signal networks and regulating mechanisms of the unfolded protein response.J Zhejiang Univ Sci B. 2017 Jan.;18(1):1-14. doi: 10.1631/jzus.B1600043. J Zhejiang Univ Sci B. 2017. PMID: 28070992 Free PMC article. Review.
-
Endoplasmic reticulum stress sensing in the unfolded protein response.Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a013169. doi: 10.1101/cshperspect.a013169. Cold Spring Harb Perspect Biol. 2013. PMID: 23388626 Free PMC article. Review.
Cited by
-
Osteogenesis imperfecta type 10 and the cellular scaffolds underlying common immunological diseases.Genes Immun. 2024 Aug;25(4):265-276. doi: 10.1038/s41435-024-00277-4. Epub 2024 May 29. Genes Immun. 2024. PMID: 38811682 Review.
References
-
- Pantazi E., Zaouali M.A., Bejaoui M., Serafin A., Folch-Puy E., Petegnief V., De Vera N., Ben Abdennebi H., Rimola A., Rosello-Catafau J. Silent Information Regulator 1 Protects the Liver against Ischemia-Reperfusion Injury: Implications in Steatotic Liver Ischemic Preconditioning. Mol. Neurobiol. 2014;27:493–503. doi: 10.1111/tri.12276. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

