Brain-Region-Specific Genes Form the Major Pathways Featuring Their Basic Functional Role: Their Implication in Animal Chronic Stress Model
- PMID: 38474132
- PMCID: PMC10931827
- DOI: 10.3390/ijms25052882
Brain-Region-Specific Genes Form the Major Pathways Featuring Their Basic Functional Role: Their Implication in Animal Chronic Stress Model
Abstract
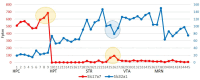
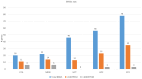
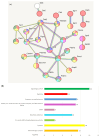
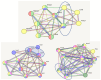
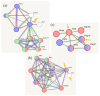
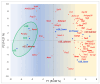
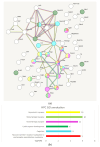
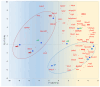
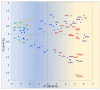
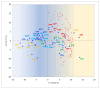
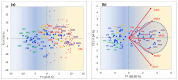
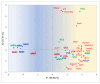
The analysis of RNA-Sec data from murine bulk tissue samples taken from five brain regions associated with behavior and stress response was conducted. The focus was on the most contrasting brain region-specific genes (BRSG) sets in terms of their expression rates. These BRSGs are identified as genes with a distinct outlying (high) expression rate in a specific region compared to others used in the study. The analysis suggested that BRSG sets form non-randomly connected compact gene networks, which correspond to the major neuron-mediated functional processes or pathways in each brain region. The number of BRSGs and the connection rate were found to depend on the heterogeneity and coordinated firing rate of neuron types in each brain region. The most connected pathways, along with the highest BRSG number, were observed in the Striatum, referred to as Medium Spiny Neurons (MSNs), which make up 95% of neurons and exhibit synchronous firing upon dopamine influx. However, the Ventral Tegmental Area/Medial Raphe Nucleus (VTA/MRN) regions, although primarily composed of monoaminergic neurons, do not fire synchronously, leading to a smaller BRSG number. The Hippocampus (HPC) region, on the other hand, displays significant neuronal heterogeneity, with glutamatergic neurons being the most numerous and synchronized. Interestingly, the two monoaminergic regions involved in the study displayed a common BRSG subnetwork architecture, emphasizing their proximity in terms of axonal throughput specifics and high-energy metabolism rates. This finding suggests the concerted evolution of monoaminergic neurons, leading to unique adaptations at the genic repertoire scale. With BRSG sets, we were able to highlight the contrasting features of the three groups: control, depressive, and aggressive mice in the animal chronic stress model. Specifically, we observed a decrease in serotonergic turnover in both the depressed and aggressive groups, while dopaminergic emission was high in both groups. There was also a notable absence of dopaminoceptive receptors on the postsynaptic membranes in the striatum in the depressed group. Additionally, we confirmed that neurogenesis BRSGs are specific to HPC, with the aggressive group showing attenuated neurogenesis rates compared to the control/depressive groups. We also confirmed that immune-competent cells like microglia and astrocytes play a crucial role in depressed phenotypes, including mitophagy-related gene Prkcd. Based on this analysis, we propose the use of BRSG sets as a suitable framework for evaluating case-control group-wise assessments of specific brain region gene pathway responses.
Keywords: animal chronic stress model; brain regions; dopamine; serotonin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Dopamine response gene pathways in dorsal striatum MSNs from a gene expression viewpoint: cAMP-mediated gene networks.BMC Neurosci. 2020 Mar 26;21(1):12. doi: 10.1186/s12868-020-00560-w. BMC Neurosci. 2020. PMID: 32216748 Free PMC article.
-
Blockade of calcium-permeable AMPA receptors in the lateral habenula produces increased antidepressant-like effects in unilateral 6-hydroxydopamine-lesioned rats compared to sham-lesioned rats.Neuropharmacology. 2019 Oct;157:107687. doi: 10.1016/j.neuropharm.2019.107687. Epub 2019 Jun 26. Neuropharmacology. 2019. PMID: 31251995
-
Enhanced serotonin and mesolimbic dopamine transmissions in a rat model of neuropathic pain.Neuropharmacology. 2015 Oct;97:383-93. doi: 10.1016/j.neuropharm.2015.06.003. Epub 2015 Jun 22. Neuropharmacology. 2015. PMID: 26113399
-
Ventral tegmental area dopamine revisited: effects of acute and repeated stress.Psychopharmacology (Berl). 2016 Jan;233(2):163-86. doi: 10.1007/s00213-015-4151-3. Epub 2015 Dec 17. Psychopharmacology (Berl). 2016. PMID: 26676983 Free PMC article. Review.
-
Dopamine-glutamate neuron projections to the nucleus accumbens medial shell and behavioral switching.Neurochem Int. 2019 Oct;129:104482. doi: 10.1016/j.neuint.2019.104482. Epub 2019 Jun 3. Neurochem Int. 2019. PMID: 31170424 Free PMC article. Review.
Cited by
-
The Role of Casr Inhibition-Mediated M2 Microglial Transformation in Ischemic Preconditioning Against Stroke.Curr Med Sci. 2025 Feb;45(1):82-92. doi: 10.1007/s11596-025-00003-9. Epub 2025 Feb 21. Curr Med Sci. 2025. PMID: 39982646
References
-
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. - DOI - PMC - PubMed
-
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. - DOI - PMC - PubMed
-
- Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

