Genome-Wide Identification and Characterization of the Sweet Orange (Citrus sinensis) GATA Family Reveals a Role for CsGATA12 as a Regulator of Citrus Bacterial Canker Resistance
- PMID: 38474170
- PMCID: PMC10931568
- DOI: 10.3390/ijms25052924
Genome-Wide Identification and Characterization of the Sweet Orange (Citrus sinensis) GATA Family Reveals a Role for CsGATA12 as a Regulator of Citrus Bacterial Canker Resistance
Abstract
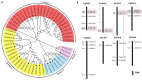
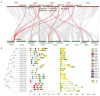
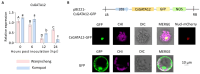
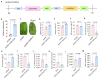
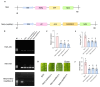
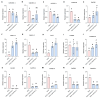
Citrus bacterial canker (CBC) is a severe bacterial infection caused by Xanthomonas citri subsp. citri (Xcc), which continues to adversely impact citrus production worldwide. Members of the GATA family are important regulators of plant development and regulate plant responses to particular stressors. This report aimed to systematically elucidate the Citrus sinensis genome to identify and annotate genes that encode GATAs and evaluate the functional importance of these CsGATAs as regulators of CBC resistance. In total, 24 CsGATAs were identified and classified into four subfamilies. Furthermore, the phylogenetic relationships, chromosomal locations, collinear relationships, gene structures, and conserved domains for each of these GATA family members were also evaluated. It was observed that Xcc infection induced some CsGATAs, among which CsGATA12 was chosen for further functional validation. CsGATA12 was found to be localized in the nucleus and was differentially upregulated in the CBC-resistant and CBC-sensitive Kumquat and Wanjincheng citrus varieties. When transiently overexpressed, CsGATA12 significantly reduced CBC resistance with a corresponding increase in abscisic acid, jasmonic acid, and antioxidant enzyme levels. These alterations were consistent with lower levels of salicylic acid, ethylene, and reactive oxygen species. Moreover, the bacteria-induced CsGATA12 gene silencing yielded the opposite phenotypic outcomes. This investigation highlights the important role of CsGATA12 in regulating CBC resistance, underscoring its potential utility as a target for breeding citrus varieties with superior phytopathogen resistance.
Keywords: GATA transcription factor; Xanthomonas citri subsp. citri (Xcc); abscisic acid (ABA); citrus bacterial canker (CBC); jasmonic acid (JA); reactive oxygen species (ROS); salicylic acid (SA).
Conflict of interest statement
The authors declare no conflicts of interests.
Figures

Similar articles
-
Genome-wide identification and characterization of the sweet orange (Citrus sinensis) basic helix-loop-helix (bHLH) family reveals a role for CsbHLH085 as a regulator of citrus bacterial canker resistance.Int J Biol Macromol. 2024 May;267(Pt 2):131442. doi: 10.1016/j.ijbiomac.2024.131442. Epub 2024 Apr 16. Int J Biol Macromol. 2024. PMID: 38621573
-
The wall-associated receptor-like kinase CsWAKL01, positively regulated by the transcription factor CsWRKY53, confers resistance to citrus bacterial canker via regulation of phytohormone signaling.J Exp Bot. 2024 Sep 27;75(18):5805-5818. doi: 10.1093/jxb/erae255. J Exp Bot. 2024. PMID: 38820225
-
Genome-wide identification and expression analyses of ABSCISIC ACID-INSENSITIVE 5 (ABI5) genes in Citrus sinensis reveal CsABI5-5 confers dual resistance to Huanglongbing and citrus canker.Int J Biol Macromol. 2025 May;306(Pt 4):141611. doi: 10.1016/j.ijbiomac.2025.141611. Epub 2025 Feb 28. Int J Biol Macromol. 2025. PMID: 40024407
-
The CsAP2-09-CsWRKY25-CsRBOH2 cascade confers resistance against citrus bacterial canker by regulating ROS homeostasis.Plant J. 2024 Apr;118(2):534-548. doi: 10.1111/tpj.16623. Epub 2024 Jan 17. Plant J. 2024. PMID: 38230828
-
Genomewide analysis of the CIII peroxidase family in sweet orange (Citrus sinensis) and expression profiles induced by Xanthomonas citri subsp. citri and hormones.J Genet. 2020;99:10. J Genet. 2020. PMID: 32089529
Cited by
-
Comparative analysis of amino acid sequence level in plant GATA transcription factors.Sci Rep. 2024 Nov 30;14(1):29786. doi: 10.1038/s41598-024-81159-7. Sci Rep. 2024. PMID: 39616200 Free PMC article.
-
GATA8-Mediated Antiviral Defence Is Countered by Tomato Chlorosis Virus-Encoded Pathogenicity Protein p27.Mol Plant Pathol. 2025 Jul;26(7):e70115. doi: 10.1111/mpp.70115. Mol Plant Pathol. 2025. PMID: 40600259 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

