Structured Tandem Repeats in Protein Interactions
- PMID: 38474241
- PMCID: PMC10931737
- DOI: 10.3390/ijms25052994
Structured Tandem Repeats in Protein Interactions
Abstract
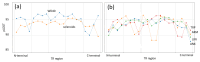
Tandem repeats (TRs) in protein sequences are consecutive, highly similar sequence motifs. Some types of TRs fold into structural units that pack together in ensembles, forming either an (open) elongated domain or a (closed) propeller, where the last unit of the ensemble packs against the first one. Here, we examine TR proteins (TRPs) to see how their sequence, structure, and evolutionary properties favor them for a function as mediators of protein interactions. Our observations suggest that TRPs bind other proteins using large, structured surfaces like globular domains; in particular, open-structured TR ensembles are favored by flexible termini and the possibility to tightly coil against their targets. While, intuitively, open ensembles of TRs seem prone to evolve due to their potential to accommodate insertions and deletions of units, these evolutionary events are unexpectedly rare, suggesting that they are advantageous for the emergence of the ancestral sequence but are early fixed. We hypothesize that their flexibility makes it easier for further proteins to adapt to interact with them, which would explain their large number of protein interactions. We provide insight into the properties of open TR ensembles, which make them scaffolds for alternative protein complexes to organize genes, RNA and proteins.
Keywords: protein evolution; protein flexibility; protein structure; protein–protein interactions; tandem repeats.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

