The Wnt Co-Receptor PTK7/Otk and Its Homolog Otk-2 in Neurogenesis and Patterning
- PMID: 38474329
- PMCID: PMC10930971
- DOI: 10.3390/cells13050365
The Wnt Co-Receptor PTK7/Otk and Its Homolog Otk-2 in Neurogenesis and Patterning
Abstract
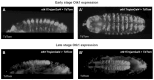
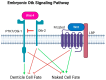
Wnt signaling is a highly conserved metazoan pathway that plays a crucial role in cell fate determination and morphogenesis during development. Wnt ligands can induce disparate cellular responses. The exact mechanism behind these different outcomes is not fully understood but may be due to interactions with different receptors on the cell membrane. PTK7/Otk is a transmembrane receptor that is implicated in various developmental and physiological processes including cell polarity, cell migration, and invasion. Here, we examine two roles of Otk-1 and Otk-2 in patterning and neurogenesis. We find that Otk-1 is a positive regulator of signaling and Otk-2 functions as its inhibitor. We propose that PTK7/Otk functions in signaling, cell migration, and polarity contributing to the diversity of cellular responses seen in Wnt-mediated processes.
Keywords: Drosophila; PTK7; Wnt.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling.EMBO J. 2011 Jul 19;30(18):3729-40. doi: 10.1038/emboj.2011.236. EMBO J. 2011. PMID: 21772251 Free PMC article.
-
The PTK7-related transmembrane proteins off-track and off-track 2 are co-receptors for Drosophila Wnt2 required for male fertility.PLoS Genet. 2014 Jul 10;10(7):e1004443. doi: 10.1371/journal.pgen.1004443. eCollection 2014 Jul. PLoS Genet. 2014. PMID: 25010066 Free PMC article.
-
Ptk7 promotes non-canonical Wnt/PCP-mediated morphogenesis and inhibits Wnt/β-catenin-dependent cell fate decisions during vertebrate development.Development. 2013 Apr;140(8):1807-18. doi: 10.1242/dev.090183. Development. 2013. PMID: 23533179
-
The many roles of PTK7: a versatile regulator of cell-cell communication.Arch Biochem Biophys. 2012 Aug 1;524(1):71-6. doi: 10.1016/j.abb.2011.12.019. Epub 2012 Jan 3. Arch Biochem Biophys. 2012. PMID: 22230326 Review.
-
The Wnt cries many: Wnt regulation of neurogenesis through tissue patterning, proliferation, and asymmetric cell division.Dev Neurobiol. 2014 Aug;74(8):772-80. doi: 10.1002/dneu.22168. Epub 2014 Mar 2. Dev Neurobiol. 2014. PMID: 24488703 Review.
Cited by
-
Unlocking Hope: Therapeutic Advances and Approaches in Modulating the Wnt Pathway for Neurodegenerative Diseases.Mol Neurobiol. 2025 Mar;62(3):3630-3652. doi: 10.1007/s12035-024-04462-4. Epub 2024 Sep 23. Mol Neurobiol. 2025. PMID: 39313658 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

