Emerging Roles and Mechanisms of RNA Modifications in Neurodegenerative Diseases and Glioma
- PMID: 38474421
- PMCID: PMC10931090
- DOI: 10.3390/cells13050457
Emerging Roles and Mechanisms of RNA Modifications in Neurodegenerative Diseases and Glioma
Abstract
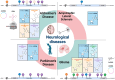
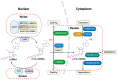
Despite a long history of research, neurodegenerative diseases and malignant brain tumor gliomas are both considered incurable, facing challenges in the development of treatments. Recent evidence suggests that RNA modifications, previously considered as static components of intracellular RNAs, are in fact dynamically regulated across various RNA species in cells and play a critical role in major biological processes in the nervous system. Innovations in next-generation sequencing have enabled the accurate detection of modifications on bases and sugars within various RNA molecules. These RNA modifications influence the stability and transportation of RNA, and crucially affect its translation. This review delves into existing knowledge on RNA modifications to offer a comprehensive inventory of these modifications across different RNA species. The detailed regulatory functions and roles of RNA modifications within the nervous system are discussed with a focus on neurodegenerative diseases and gliomas. This article presents a comprehensive overview of the fundamental mechanisms and emerging roles of RNA modifications in these diseases, which can facilitate the creation of innovative diagnostics and therapeutics for these conditions.
Keywords: RNA modifications; glioma; neurodegeneration.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Regulation and roles of RNA modifications in aging-related diseases.Aging Cell. 2022 Jul;21(7):e13657. doi: 10.1111/acel.13657. Epub 2022 Jun 19. Aging Cell. 2022. PMID: 35718942 Free PMC article. Review.
-
RNA-Guided RNA Modifications: Biogenesis, Functions, and Applications.Acc Chem Res. 2023 Nov 21;56(22):3198-3210. doi: 10.1021/acs.accounts.3c00474. Epub 2023 Nov 6. Acc Chem Res. 2023. PMID: 37931323
-
Epigenomic Reprogramming as a Driver of Malignant Glioma.Cancer Cell. 2020 Nov 9;38(5):647-660. doi: 10.1016/j.ccell.2020.08.008. Epub 2020 Sep 10. Cancer Cell. 2020. PMID: 32916125 Free PMC article. Review.
-
Circular RNAs: Functions and Prospects in Glioma.J Mol Neurosci. 2019 Jan;67(1):72-81. doi: 10.1007/s12031-018-1211-2. Epub 2018 Nov 20. J Mol Neurosci. 2019. PMID: 30460608 Review.
-
RNA Modifications and RNA Metabolism in Neurological Disease Pathogenesis.Int J Mol Sci. 2021 Nov 1;22(21):11870. doi: 10.3390/ijms222111870. Int J Mol Sci. 2021. PMID: 34769301 Free PMC article. Review.
Cited by
-
The dual mechanism of m6A demethylase ALKBH5 in regulating energy metabolism during exposure to MC-LR.Cell Death Dis. 2025 Jul 3;16(1):489. doi: 10.1038/s41419-025-07791-x. Cell Death Dis. 2025. PMID: 40610445 Free PMC article.
-
Identification of deregulated lncRNAs in Alzheimer's disease: an integrated gene co-expression network analysis of hippocampus and fusiform gyrus RNA-seq datasets.Front Aging Neurosci. 2024 Jul 17;16:1437278. doi: 10.3389/fnagi.2024.1437278. eCollection 2024. Front Aging Neurosci. 2024. PMID: 39086756 Free PMC article.
-
Epigenetic Mechanisms in Aging: Extrinsic Factors and Gut Microbiome.Genes (Basel). 2024 Dec 14;15(12):1599. doi: 10.3390/genes15121599. Genes (Basel). 2024. PMID: 39766866 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

