Identification of Phytochemicals from Arabian Peninsula Medicinal Plants as Strong Binders to SARS-CoV-2 Proteases (3CLPro and PLPro) by Molecular Docking and Dynamic Simulation Studies
- PMID: 38474509
- PMCID: PMC10934083
- DOI: 10.3390/molecules29050998
Identification of Phytochemicals from Arabian Peninsula Medicinal Plants as Strong Binders to SARS-CoV-2 Proteases (3CLPro and PLPro) by Molecular Docking and Dynamic Simulation Studies
Abstract
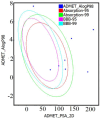
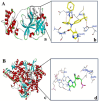
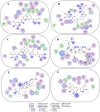
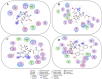
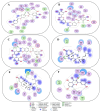
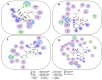
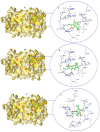
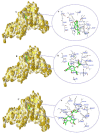
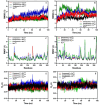
We provide promising computational (in silico) data on phytochemicals (compounds 1-10) from Arabian Peninsula medicinal plants as strong binders, targeting 3-chymotrypsin-like protease (3CLPro) and papain-like proteases (PLPro) of SARS-CoV-2. Compounds 1-10 followed the Lipinski rules of five (RO5) and ADMET analysis, exhibiting drug-like characters. Non-covalent (reversible) docking of compounds 1-10 demonstrated their binding with the catalytic dyad (CYS145 and HIS41) of 3CLPro and catalytic triad (CYS111, HIS272, and ASP286) of PLPro. Moreover, the implementation of the covalent (irreversible) docking protocol revealed that only compounds 7, 8, and 9 possess covalent warheads, which allowed the formation of the covalent bond with the catalytic dyad (CYS145) in 3CLPro and the catalytic triad (CYS111) in PLPro. Root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and radius of gyration (Rg) analysis from molecular dynamic (MD) simulations revealed that complexation between ligands (compounds 7, 8, and 9) and 3CLPro and PLPro was stable, and there was less deviation of ligands. Overall, the in silico data on the inherent properties of the above phytochemicals unravel the fact that they can act as reversible inhibitors for 3CLPro and PLPro. Moreover, compounds 7, 8, and 9 also showed their novel properties to inhibit dual targets by irreversible inhibition, indicating their effectiveness for possibly developing future drugs against SARS-CoV-2. Nonetheless, to confirm the theoretical findings here, the effectiveness of the above compounds as inhibitors of 3CLPro and PLPro warrants future investigations using suitable in vitro and in vivo tests.
Keywords: 3CLPro; PLPro; SARS-CoV-2; docking; medicinal plants; molecular dynamic simulations.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- WHO WHO Coronavirus (COVID-19) Dashboard. 2023. [(accessed on 22 November 2023)]. Available online: https://covid19.who.int.
-
- Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z., Liu X., Wei L., Truelove S.A., Zhang T., et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: A retrospective cohort study. Lancet Infect. Dis. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. - DOI - PMC - PubMed
-
- Amaral-Machado L., Oliveira W.N., Rodrigues V.M., Albuquerque N.A., Alencar É.N., Egito E.S.T. Could natural products modulate early inflammatory responses, preventing acute respiratory distress syndrome in COVID-19-confirmed patients? Biomed. Pharmacother. 2021;134:111143. doi: 10.1016/j.biopha.2020.111143. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

