Functional Characterization of the MsFKF1 Gene Reveals Its Dual Role in Regulating the Flowering Time and Plant Height in Medicago sativa L
- PMID: 38475501
- PMCID: PMC10934225
- DOI: 10.3390/plants13050655
Functional Characterization of the MsFKF1 Gene Reveals Its Dual Role in Regulating the Flowering Time and Plant Height in Medicago sativa L
Abstract
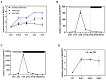
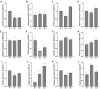
Alfalfa (M. sativa), a perennial legume forage, is known for its high yield and good quality. As a long-day plant, it is sensitive to changes in the day length, which affects the flowering time and plant growth, and limits alfalfa yield. Photoperiod-mediated delayed flowering in alfalfa helps to extend the vegetative growth period and increase the yield. We isolated a blue-light phytohormone gene from the alfalfa genome that is an ortholog of soybean FKF1 and named it MsFKF1. Gene expression analyses showed that MsFKF1 responds to blue light and the circadian clock in alfalfa. We found that MsFKF1 regulates the flowering time through the plant circadian clock pathway by inhibiting the transcription of E1 and COL, thus suppressing FLOWERING LOCUS T a1 (FTa1) transcription. In addition, transgenic lines exhibited higher plant height and accumulated more biomass in comparison to wild-type plants. However, the increased fiber (NDF and ADF) and lignin content also led to a reduction in the digestibility of the forage. The key genes related to GA biosynthesis, GA20OX1, increased in the transgenic lines, while GA2OX1 decreased for the inactive GA transformation. These findings offer novel insights on the function of MsFKF1 in the regulation of the flowering time and plant height in cultivated M. sativa. These insights into MsFKF1's roles in alfalfa offer potential strategies for molecular breeding aimed at optimizing flowering time and biomass yield.
Keywords: FKF1; alfalfa; blue light; circadian clock; flowering time.
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Figures




Similar articles
-
Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality.Plant Biotechnol J. 2023 Jul;21(7):1383-1392. doi: 10.1111/pbi.14042. Epub 2023 Mar 25. Plant Biotechnol J. 2023. PMID: 36964962 Free PMC article.
-
Improvement of alfalfa forage quality and management through the down-regulation of MsFTa1.Plant Biotechnol J. 2020 Apr;18(4):944-954. doi: 10.1111/pbi.13258. Epub 2019 Oct 13. Plant Biotechnol J. 2020. PMID: 31536663 Free PMC article.
-
Shade Delayed Flowering Phenology and Decreased Reproductive Growth of Medicago sativa L.Front Plant Sci. 2022 Jun 2;13:835380. doi: 10.3389/fpls.2022.835380. eCollection 2022. Front Plant Sci. 2022. PMID: 35720597 Free PMC article.
-
Seasonal and Diurnal Transcriptome Atlas in Natural Environment Reveals Flowering Time Regulatory Network in Alfalfa.Plant Cell Environ. 2025 Jul;48(7):4723-4739. doi: 10.1111/pce.15466. Epub 2025 Mar 12. Plant Cell Environ. 2025. PMID: 40070311
-
Molecular mechanisms for the photoperiodic regulation of flowering in soybean.J Integr Plant Biol. 2021 Jun;63(6):981-994. doi: 10.1111/jipb.13021. Epub 2021 Apr 26. J Integr Plant Biol. 2021. PMID: 33090664 Review.
Cited by
-
Genome-wide identification and expression analysis of CBF/DREB1 gene family in Medicago sativa L. and functional verification of MsCBF9 affecting flowering time.BMC Plant Biol. 2025 Jan 22;25(1):87. doi: 10.1186/s12870-025-06081-0. BMC Plant Biol. 2025. PMID: 39838277 Free PMC article.
-
Advances in basic biology of alfalfa (Medicago sativa L.): a comprehensive overview.Hortic Res. 2025 Mar 10;12(7):uhaf081. doi: 10.1093/hr/uhaf081. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40343348 Free PMC article.
-
Engineering plant photoreceptors towards enhancing plant productivity.Plant Mol Biol. 2025 May 6;115(3):64. doi: 10.1007/s11103-025-01591-9. Plant Mol Biol. 2025. PMID: 40327169 Review.
References
-
- Wolabu T.W., Mahmood K., Jerez I.T., Cong L., Yun J., Udvardi M., Tadege M., Wang Z., Wen J. Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 2023;21:1383–1392. doi: 10.1111/pbi.14042. - DOI - PMC - PubMed
-
- Bao Q., Wolabu T.W., Zhang Q., Zhang T., Liu Z., Sun J., Wang Z.-Y. Application of CRISPR/Cas9 technology in forages. Grassland Res. 2022;1:244–251. doi: 10.1002/glr2.12036. - DOI
-
- Chen H., Zeng Y., Yang Y., Huang L., Tang B., Zhang H., Hao F., Liu W., Li Y., Liu Y., et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020;11:2494. doi: 10.1038/s41467-020-16338-x. - DOI - PMC - PubMed
-
- Lorenzo C.D., García-Gagliardi P., Antonietti M.S., Sánchez-Lamas M., Mancini E., Dezar C.A., Vazquez M., Watson G., Yanovsky M.J., Cerdán P.D. Improvement of alfalfa forage quality and management through the down-regulation of MsFTa1. Plant Biotechnol. J. 2020;18:944–954. doi: 10.1111/pbi.13258. - DOI - PMC - PubMed
-
- Zhou C., Han L., Pislariu C., Nakashima J., Fu C., Jiang Q., Quan L., Blancaflor E.B., Tang Y., Bouton J.H., et al. From model to crop: Functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011;157:1483–1496. doi: 10.1104/pp.111.185140. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

