Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.)
- PMID: 38475522
- PMCID: PMC10935285
- DOI: 10.3390/plants13050676
Identification of WRKY Family Members and Characterization of the Low-Temperature-Stress-Responsive WRKY Genes in Luffa (Luffa cylindrica L.)
Abstract
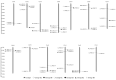
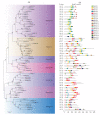
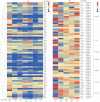
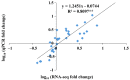
The plant-specific WRKY transcription factor family members have diverse regulatory effects on the genes associated with many plant processes. Although the WRKY proteins in Arabidopsis thaliana and other species have been thoroughly investigated, there has been relatively little research on the WRKY family in Luffa cylindrica, which is one of the most widely grown vegetables in China. In this study, we performed a genome-wide analysis to identify L. cylindrica WRKY genes, which were subsequently classified and examined in terms of their gene structures, chromosomal locations, promoter cis-acting elements, and responses to abiotic stress. A total of 62 LcWRKY genes (471-2238 bp) were identified and divided into three phylogenetic groups (I, II, and III), with group II further divided into five subgroups (IIa, IIb, IIc, IId, and IIe) in accordance with the classification in other plants. The LcWRKY genes were unevenly distributed across 13 chromosomes. The gene structure analysis indicated that the LcWRKY genes contained 0-11 introns (average of 4.4). Moreover, 20 motifs were detected in the LcWRKY proteins with conserved motifs among the different phylogenetic groups. Two subgroup IIc members (LcWRKY16 and LcWRKY31) contained the WRKY sequence variant WRKYGKK. Additionally, nine cis-acting elements related to diverse responses to environmental stimuli were identified in the LcWRKY promoters. The subcellular localization analysis indicated that three LcWRKY proteins (LcWRKY43, LcWRKY7, and LcWRKY23) are localized in the nucleus. The tissue-specific LcWRKY expression profiles reflected the diversity in LcWRKY expression. The RNA-seq data revealed the effects of low-temperature stress on LcWRKY expression. The cold-induced changes in expression were verified via a qRT-PCR analysis of 24 differentially expressed WRKY genes. Both LcWRKY7 and LcWRKY12 were highly responsive to the low-temperature treatment (approximately 110-fold increase in expression). Furthermore, the LcWRKY8, LcWRKY12, and LcWRKY59 expression levels increased by more than 25-fold under cold conditions. Our findings will help clarify the evolution of the luffa WRKY family while also providing valuable insights for future studies on WRKY functions.
Keywords: Luffa cylindrica; WRKY transcription factors; abiotic stress; expression analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources

