Allelic variation in the autotetraploid potato: genes involved in starch and steroidal glycoalkaloid metabolism as a case study
- PMID: 38475714
- PMCID: PMC10936075
- DOI: 10.1186/s12864-024-10186-5
Allelic variation in the autotetraploid potato: genes involved in starch and steroidal glycoalkaloid metabolism as a case study
Abstract
Background: Tuber starch and steroidal glycoalkaloid (SGA)-related traits have been consistently prioritized in potato breeding, while allelic variation pattern of genes that underlie these traits is less explored.
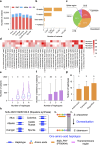
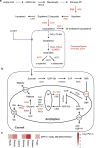
Results: Here, we focused on the genes involved in two important metabolic pathways in the potato: starch metabolism and SGA biosynthesis. We identified 119 genes consisting of 81 involved in starch metabolism and 38 in the biosynthesis of steroidal glycoalkaloids, and discovered 96,166 allelic variants among 2,169 gene haplotypes in six autotetraploid potato genomes. Comparative analyses revealed an uneven distribution of allelic variants among gene haplotypes and that the vast majority of deleterious mutations in these genes are retained in heterozygous state in the autotetraploid potato genomes. Leveraging full-length cDNA sequencing data, we find that approximately 70% of haplotypes of the 119 genes are transcribable. Population genetic analyses identify starch and SGA biosynthetic genes that are potentially conserved or diverged between potato varieties with varying starch or SGA content.
Conclusions: These results deepen the understanding of haplotypic diversity within functionally important genes in autotetraploid genomes and may facilitate functional characterization of genes or haplotypes contributing to traits related to starch and SGA in potato.
Keywords: Allelic variation; Potato; Starch; Steroidal glycoalkaloid.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Natural diversity of potato (Solanum tuberosum) invertases.BMC Plant Biol. 2010 Dec 9;10:271. doi: 10.1186/1471-2229-10-271. BMC Plant Biol. 2010. PMID: 21143910 Free PMC article.
-
Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L.) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array.BMC Genomics. 2017 Aug 22;18(1):642. doi: 10.1186/s12864-017-3979-9. BMC Genomics. 2017. PMID: 28830357 Free PMC article.
-
Allelic variation in genes contributing to glycoalkaloid biosynthesis in a diploid interspecific population of potato.Theor Appl Genet. 2014 Feb;127(2):391-405. doi: 10.1007/s00122-013-2226-2. Epub 2013 Nov 5. Theor Appl Genet. 2014. PMID: 24190104
-
Gene silencing in potato: allelic differences and effect of ploidy.Plant Mol Biol. 2000 Jun;43(2-3):377-86. doi: 10.1023/a:1006476621946. Plant Mol Biol. 2000. PMID: 10999417 Review.
-
Potato improvement through genetic engineering.GM Crops Food. 2021 Jan 2;12(1):479-496. doi: 10.1080/21645698.2021.1993688. GM Crops Food. 2021. PMID: 34991415 Free PMC article. Review.
Cited by
-
Potato steroidal glycoalkaloids: properties, biosynthesis, regulation and genetic manipulation.Mol Hortic. 2024 Dec 13;4(1):43. doi: 10.1186/s43897-024-00118-y. Mol Hortic. 2024. PMID: 39668379 Free PMC article. Review.
-
SpudDB: a database for accessing potato genomic data.Genetics. 2025 Mar 17;229(3):iyae205. doi: 10.1093/genetics/iyae205. Genetics. 2025. PMID: 39657689 Free PMC article.
References
-
- Wijesinha-Bettoni R, Mouillé B. The contribution of potatoes to global food security, nutrition and healthy diets. Am J Potato Res. 2019;96(2):139–49. doi: 10.1007/s12230-018-09697-1. - DOI
-
- Devaux A, Kromann P, Ortiz O. Potatoes for sustainable global food security. Potato Res. 2014;57(3):185–99. doi: 10.1007/s11540-014-9265-1. - DOI
-
- Schönhals EM, Ortega F, Barandalla L, Aragones A, Ruiz de Galarreta JI, Liao JC, Sanetomo R, Walkemeier B, Tacke E, Ritter E, et al. Identification and reproducibility of diagnostic DNA markers for tuber starch and yield optimization in a novel association mapping population of potato (Solanum tuberosum L) Theor Appl Genet. 2016;129(4):767–85. doi: 10.1007/s00122-016-2665-7. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

