Cellular and clinical impact of protein phosphatase enzyme epigenetic silencing in multiple cancer tissues
- PMID: 38475971
- PMCID: PMC10935810
- DOI: 10.1186/s40246-024-00592-x
Cellular and clinical impact of protein phosphatase enzyme epigenetic silencing in multiple cancer tissues
Abstract
Background: Protein Phosphatase Enzymes (PPE) and protein kinases simultaneously control phosphorylation mechanisms that tightly regulate intracellular signalling pathways and stimulate cellular responses. In human malignancies, PPE and protein kinases are frequently mutated resulting in uncontrolled kinase activity and PPE suppression, leading to cell proliferation, migration and resistance to anti-cancer therapies. Cancer associated DNA hypermethylation at PPE promoters gives rise to transcriptional silencing (epimutations) and is a hallmark of cancer. Despite recent advances in sequencing technologies, data availability and computational capabilities, only a fraction of PPE have been reported as transcriptionally inactive as a consequence of epimutations.
Methods: In this study, we examined promoter-associated DNA methylation profiles in Protein Phosphatase Enzymes and their Interacting Proteins (PPEIP) in a cohort of 705 cancer patients in five tissues (Large intestine, Oesophagus, Lung, Pancreas and Stomach) in three cell models (primary tumours, cancer cell lines and 3D embedded cancer cell cultures). As a subset of PPEIP are known tumour suppressor genes, we analysed the impact of PPEIP promoter hypermethylation marks on gene expression, cellular networks and in a clinical setting.
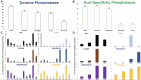
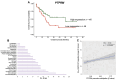
Results: Here, we report epimutations in PPEIP are a frequent occurrence in the cancer genome and manifest independent of transcriptional activity. We observed that different tumours have varying susceptibility to epimutations and identify specific cellular signalling networks that are primarily affected by epimutations. Additionally, RNA-seq analysis showed the negative impact of epimutations on most (not all) Protein Tyrosine Phosphatase transcription. Finally, we detected novel clinical biomarkers that inform on patient mortality and anti-cancer treatment sensitivity.
Conclusions: We propose that DNA hypermethylation marks at PPEIP frequently contribute to the pathogenesis of malignancies and within the precision medicine space, hold promise as biomarkers to inform on clinical features such as patient survival and therapeutic response.
Keywords: Biomarker; Cancer; DNA methylation; Epigenetics; Gene-silencing; Hyper-methylation; Protein prosphatase enzymes; RNA-seq; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
An integrated epigenomic-transcriptomic landscape of lung cancer reveals novel methylation driver genes of diagnostic and therapeutic relevance.Theranostics. 2021 Mar 11;11(11):5346-5364. doi: 10.7150/thno.58385. eCollection 2021. Theranostics. 2021. PMID: 33859751 Free PMC article.
-
ROR2 is epigenetically inactivated in the early stages of colorectal neoplasia and is associated with proliferation and migration.BMC Cancer. 2016 Jul 20;16:508. doi: 10.1186/s12885-016-2576-7. BMC Cancer. 2016. PMID: 27440078 Free PMC article.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
Epigenetic silencing of TGFBI confers resistance to trastuzumab in human breast cancer.Breast Cancer Res. 2019 Jul 5;21(1):79. doi: 10.1186/s13058-019-1160-x. Breast Cancer Res. 2019. PMID: 31277676 Free PMC article.
-
[Germ-line epimutations and human cancer].Ai Zheng. 2009 Dec;28(12):1236-42. doi: 10.5732/cjc.009.10266. Ai Zheng. 2009. PMID: 19958615 Review. Chinese.
Cited by
-
Distinctive chromosomal, mutational and transcriptional profiling in colon versus rectal cancers.J Transl Med. 2025 Aug 6;23(1):869. doi: 10.1186/s12967-025-06908-2. J Transl Med. 2025. PMID: 40770356 Free PMC article.
References
-
- Li X, Wilmanns M, Thornton J, Köhn M. Elucidating human phosphatase-substrate networks. Sci Signal. 2013;6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

