Nucleation of Huntingtin Aggregation Proceeds via Conformational Conversion of Pre-Formed, Sparsely-Populated Tetramers
- PMID: 38476051
- PMCID: PMC11199967
- DOI: 10.1002/advs.202309217
Nucleation of Huntingtin Aggregation Proceeds via Conformational Conversion of Pre-Formed, Sparsely-Populated Tetramers
Abstract
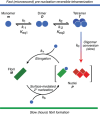
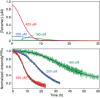
Pathogenic huntingtin exon-1 protein (httex1), characterized by an expanded polyglutamine tract located between the N-terminal amphiphilic region and a C-terminal polyproline-rich domain, forms fibrils that accumulate in neuronal inclusion bodies, and is associated with a fatal, autosomal dominant neurodegenerative condition known as Huntington's disease. Here a complete kinetic model is described for aggregation/fibril formation of a httex1 construct with a 35-residue polyglutamine repeat, httex1Q35. Using exchange NMR spectroscopy, it is previously shown that the reversible formation of a sparsely-populated tetramer of the N-terminal amphiphilic domain of httex1Q35, comprising a D2 symmetric four-helix bundle, occurs on the microsecond time-scale and is a prerequisite for subsequent nucleation and fibril formation on a time scale that is many orders of magnitude slower (hours). Here a unified kinetic model of httex1Q35 aggregation is developed in which fast, reversible tetramerization is directly linked to slow irreversible fibril formation via conversion of pre-equilibrated tetrameric species to "active", chain elongation-capable nuclei by conformational re-arrangement with a finite, monomer-independent rate. The unified model permits global quantitative analysis of reversible tetramerization and irreversible fibril formation from a time series of 1H-15N correlation spectra recorded during the course of httex1Q35 aggregation.
Keywords: NMR; elongation competent nuclei; huntingtin exon‐1 protein; pre‐nucleation tetramers; unified kinetic model of aggregation.
Published 2024. This article is a U.S. Government work and is in the public domain in the USA. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Effects of Macromolecular Cosolutes on the Kinetics of Huntingtin Aggregation Monitored by NMR Spectroscopy.J Phys Chem Lett. 2024 Jun 20;15(24):6375-6382. doi: 10.1021/acs.jpclett.4c01410. Epub 2024 Jun 10. J Phys Chem Lett. 2024. PMID: 38857530 Free PMC article.
-
Quantitative NMR analysis of the kinetics of prenucleation oligomerization and aggregation of pathogenic huntingtin exon-1 protein.Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2207690119. doi: 10.1073/pnas.2207690119. Epub 2022 Jul 12. Proc Natl Acad Sci U S A. 2022. PMID: 35858329 Free PMC article.
-
Quantitative Exchange NMR-Based Analysis of Huntingtin-SH3 Interactions Suggests an Allosteric Mechanism of Inhibition of Huntingtin Aggregation.J Am Chem Soc. 2021 Jun 30;143(25):9672-9681. doi: 10.1021/jacs.1c04786. Epub 2021 Jun 17. J Am Chem Soc. 2021. PMID: 34137596 Free PMC article.
-
Proteostasis of Huntingtin in Health and Disease.Int J Mol Sci. 2017 Jul 19;18(7):1568. doi: 10.3390/ijms18071568. Int J Mol Sci. 2017. PMID: 28753941 Free PMC article. Review.
-
The pathobiology of perturbed mutant huntingtin protein-protein interactions in Huntington's disease.J Neurochem. 2019 Nov;151(4):507-519. doi: 10.1111/jnc.14853. Epub 2019 Sep 15. J Neurochem. 2019. PMID: 31418858 Review.
Cited by
-
Effects of Macromolecular Cosolutes on the Kinetics of Huntingtin Aggregation Monitored by NMR Spectroscopy.J Phys Chem Lett. 2024 Jun 20;15(24):6375-6382. doi: 10.1021/acs.jpclett.4c01410. Epub 2024 Jun 10. J Phys Chem Lett. 2024. PMID: 38857530 Free PMC article.
-
Direct Observation of Secondary Nucleation in Huntingtin Amyloid Formation by High-Speed Atomic Force Microscopy.J Am Chem Soc. 2025 Jun 25;147(25):21973-21984. doi: 10.1021/jacs.5c05571. Epub 2025 Jun 12. J Am Chem Soc. 2025. PMID: 40505012 Free PMC article.
-
Characterizing heterogeneity in amyloid formation processes.Curr Opin Struct Biol. 2024 Dec;89:102951. doi: 10.1016/j.sbi.2024.102951. Epub 2024 Nov 19. Curr Opin Struct Biol. 2024. PMID: 39566372 Review.
-
Modeling Protein Aggregation Kinetics from NMR Data.J Mol Biol. 2025 Jun 9:169269. doi: 10.1016/j.jmb.2025.169269. Online ahead of print. J Mol Biol. 2025. PMID: 40499748 Review.
References
-
- Bates G. P., Dorsey R., Gusella J. F., Hayden M. R., Kay C., Leavitt B. R., Nance M., Ross C. A., Scahill R. I., Wetzel R., Wild E. J., Tabrizi S. J., Nat. Rev. Dis. Primers 2015, 1, 15005. - PubMed
-
- Andresen J. M., Gayán J., Djoussé L., Roberts S., Brocklebank D., Cherny S. S., Ann. Hum. Genet. 2007, 71, 295. - PubMed
-
- Ross C. A., Tabrizi S. J., Lancet Neurol. 2011, 10, 83. - PubMed
-
- Zuccato C., Valenza M., Cattaneo E., Physiol. Rev. 2010, 90, 905. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
