Functional support vector machine
- PMID: 38476094
- PMCID: PMC11639177
- DOI: 10.1093/biostatistics/kxae007
Functional support vector machine
Erratum in
-
Correction.Biostatistics. 2024 Dec 31;26(1):kxae029. doi: 10.1093/biostatistics/kxae029. Biostatistics. 2024. PMID: 39186534 Free PMC article. No abstract available.
Abstract
Linear and generalized linear scalar-on-function modeling have been commonly used to understand the relationship between a scalar response variable (e.g. continuous, binary outcomes) and functional predictors. Such techniques are sensitive to model misspecification when the relationship between the response variable and the functional predictors is complex. On the other hand, support vector machines (SVMs) are among the most robust prediction models but do not take account of the high correlations between repeated measurements and cannot be used for irregular data. In this work, we propose a novel method to integrate functional principal component analysis with SVM techniques for classification and regression to account for the continuous nature of functional data and the nonlinear relationship between the scalar response variable and the functional predictors. We demonstrate the performance of our method through extensive simulation experiments and two real data applications: the classification of alcoholics using electroencephalography signals and the prediction of glucobrassicin concentration using near-infrared reflectance spectroscopy. Our methods especially have more advantages when the measurement errors in functional predictors are relatively large.
Keywords: EEG; functional data analysis; functional principal component analysis; scalar-on-function modeling; support vector machine.
© The Author 2024. Published by Oxford University Press. All rights reserved. For Permissions, email: journals.permissions@oup.com.
Conflict of interest statement
None declared.
Figures
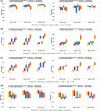

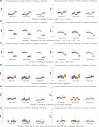


References
-
- Berlinet A, Thomas-Agnan C. Reproducing kernel Hilbert spaces in probability and statistics. New York, NY: Springer Science & Business Media; 2011.
-
- Besse P, Ramsay JO. Principal components analysis of sampled functions. Psychometrika. 1986:51:285–311.
-
- Bishop CM, Nasrabadi NM. Pattern recognition and machine learning. vol. 4. New York, NY: Springer; 2006.
-
- Caputo B, Sim K, Furesjo F, Smola A. Appearance-based object recognition using SVMs: which kernel should I use? In Proceedings of NIPS workshop on Statistical methods for computational experiments in visual processing and computer vision, Whistler. 2002.
-
- Font R, Del Rio-Celestino M, Rosa E, Aires A, De Haro-Bailon A. Glucosinolate assessment in brassica oleracea leaves by near-infrared spectroscopy. J Agric Sci. 2005:143(1):65–73.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

